Life from Scratch
Learning to make synthetic cells
Maggots don’t arise spontaneously out of dead, rotting meat. Aphids never materialize within drops of morning dew. Aristotle and others who believed in the spontaneous generation of life were dead wrong.
The only time life arose from nonlife, biologists believe, was almost 4 billion years ago, when simple living cells first appeared on Earth. But now, with the help of a microscopic parasite that infects people’s genitals, researchers may rehabilitate the core of Aristotle’s idea.
Scientists are on the verge of creating living cells by piecing together small molecules that are themselves not alive. The result would be the world’s first human-made life forms, synthetic cells made more or less from scratch.
The goal is to make cells that are as simple as possible, yet still able to grow, reproduce, and evolve—some of the defining traits of life.
“Simplicity has always been where we try to gain understanding,” says John Glass of the J. Craig Venter Institute in Rockville, Md. “In a way, what we’re doing is making a better platform for understanding what life is.” It’s a bit like learning the essentials of how a luxury car works by building a dune buggy from spare parts.
Some scientists, including Glass, hope to make such a minimal cell by whittling down the genome of an existing bacterium to its barest elements, and then synthesizing that minimal genome. In the lab, scientists can assemble the genomic DNA by piecing together chemicals called nucleotides, which constitute the individual letters of genetic code.
Other scientists, starting from long lists of molecules and genes, are devising plans to assemble these parts to make an entire cell, not just its genome, by hand.
Still other researchers take a radically different approach. Instead of trying to construct cells from the same proteins and DNA found in modern organisms, these investigators hope to assemble a cell from more-primitive molecules that better mimic the molecules probably involved in the origin of life. If successful, these scientists may uncover clues about how the original “spontaneous generation” of life occurred billions of years ago.
The first step, however, involves that irritating parasite.
Starting small
Being a parasite has its perks. Living inside a mammalian host, a microbe enjoys a constant, comfy temperature and an endless supply of nutrients.
Because their living conditions are relatively cozy, parasitic microbes can get by with much smaller genetic toolkits. That’s why one of the smallest known bacterial genomes belongs to Mycoplasma genitalium, a parasite that can infect the cervix and vagina of women and the urinary tracts of women and men.
While the human genome contains more than 3 billion letters of DNA code, M. genitalium has only about 580,000. This tiny genome encodes a mere 528 genes.
Even that’s too many for Hamilton Smith, a Nobel laureate and Glass’ colleague at the Venter Institute. For the past 9 years, Smith’s team has been systematically removing single genes from M. genitalium to see which ones the bacterium can survive without. So far, the group has found about 100 genes that are dispensable when removed individually. Many of these genes help the microbe evade the human immune system, which isn’t necessary in lab dishes.
However, “it’s unlikely that all of the 100 could be removed simultaneously,” Smith said last June at the Synthetic Biology 3.0 conference in Zurich. Some genes perform redundant functions, so if Smith’s team removes one of these, the microbe can keep chugging along. But if the researchers take out all of the genes responsible for a crucial task, the organism will die.
With so many genes that might be expendable, testing every possible combination by hand would take years. So instead, the scientists produce thousands of combinations quickly and grow them in lab dishes to see which ones survive. “You let biological selection tell you what’s necessary,” Glass says.
Using this method, the team is homing in on a set of essential genes. With this minimal genome in hand, the researchers plan to build the DNA synthetically by stringing together individual nucleotides in the right sequence. Assembly of made-to-order DNA has become routine, but nobody has ever put together a single DNA molecule that’s hundreds of thousands of nucleotides long.
To synthesize an entire minimal genome, Smith and his colleagues are stitching together medium-length segments of DNA using enzymes to join matching, overlapping regions capping the ends of each fragment. This way, the team can assemble the genome piecewise rather than trying to synthesize it all at once.
“We eventually want to make an organism called Mycoplasma laboratorium,” Smith says. The more familiar name for this hypothetical cell is Synthia.
Glass says that the team is on the verge of making such a cell within the next few months. In the Aug. 3, 2007 Science, the researchers announced that they had transplanted the entire genome of one species of Mycoplasma into a related species. The recipient cells began using the foreign genome as if it was their own, showing that the receiving cells can “boot up” the newly inserted DNA (SN: 6/30/07, p. 403). All that remains is to finish piecing together a minimal, synthetic genome and then to insert it into a Mycoplasma bacterium by the same technique.
Beyond serving as a hobby-kit cell for unraveling basic cell biology, Synthia might serve as a platform for developing novel biotechnologies.
“Learning how to rebuild something will give us control over a cell that we don’t now have,” Glass says. Craig Venter, the scientist-cum-biotech tycoon who led the private effort to map the human genome and now heads the Venter Institute, has said he hopes that a minimal genome will serve as a base upon which to add custom functions, such as genes for converting feedstock into hydrogen for fuel.
“Ultimately I’m not going to want a cell that’s capable of a great many things,” Glass says. “I’m going to want a cell that can do one thing for me.”
This narrowness of function could help address possible safety concerns. “Mycoplasmas are extremely wimpy organisms, and minimal organisms will be even more limited,” Glass says. Because of their barebones genetic toolkits, such microbes would only survive under carefully controlled laboratory conditions.
Assembly required
Synthia would be an important scientific milestone, but it would still be only partially synthetic.
The genome would be human-made, but the membrane enclosing the cell and the complex blend of proteins in the cell body would have come from the living bacterium that received the injected genome.
“Essentially, we are commandeering the shell of a [preexisting] cell,” Glass says.
Another shortcoming of the Venter Institute’s approach, in some scientists’ view, is that the team would have created a semisynthetic cell without fully understanding their creation.
“Even in Mycoplasma, the functions of a fifth of the genes are unknown,” says Tony Forster of Vanderbilt University in Nashville. “So, the concept of using that as a minimal organism is still fraught with an enormous number of unknowns, which doesn’t really give you much understanding of what’s going on.”
Forster is one of several scientists pursuing a more comprehensive approach that could be years, instead of months, away from completion. The idea is to piece together all of the cell’s essential systems using small molecules to produce something that’s alive—if just barely.
“I realized that I had most of the parts I needed in the [laboratory] freezer, so it’s time to start thinking about putting together the whole thing,” Forster says. In theory, this bottom-up approach would allow scientists to understand all of the parts of the cells that they make.
Working with George Church of Harvard University, Forster published a detailed recipe for how to make a living cell from scratch in the August 2006 Molecular Systems Biology. Their plan calls for a synthetic genome containing 151 genes that produce all the basic molecular machinery for protein creation and DNA replication.
Wrapping this manmade genome in a spherical shell of lipid molecules—along with some chemically synthesized protein-production molecules to get protein biosynthesis started—should in theory yield a living, growing cell.
It may sound farfetched, but scientists have already taken some significant steps. Making a lipid bubble to serve as a membrane is fairly easy. Just as water beads up on a waxy surface, fat molecules in water will spontaneously cluster into thin spheres called liposomes. In fact, real cell membranes also consist of a kind of lipid. Scientists have long thought that this suggests that the first cells may have evolved from protocells formed by this natural beading up of fat molecules.
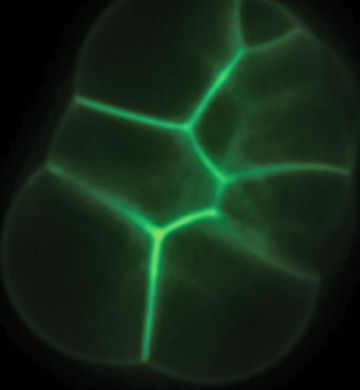
In 2004, Albert J. Libchaber of the Rockefeller University in New York City created liposomes and added the gene for a green fluorescent protein as well as all the molecular machinery needed to convert the genetic code into proteins. The result: green, glowing fat bubbles.
It was a deceivingly simple demonstration. Protein synthesis is extremely complicated, and it’s crucial for living cells because proteins do most of the work that keeps cells alive. However, Libchaber’s team used an extract from the bacterium E. coli to provide the protein-making system, essentially borrowing the entire system from a living organism without knowing fully what it consisted of or how it worked.
More recently, Giovanni Murtas of the Enrico Fermi Research Centre in Rome succeeded in making green-glowing liposomes by instead using a cocktail of 36 enzymes and other molecules that had been synthesized by a team of scientists led by Hiroshi Ueda at University of Tokyo in Japan. The researchers reported the results in the Nov. 9, 2007 Biochemical and Biophysical Research Communications.
Of course, protein-making fat bubbles still fall far short of a growing, reproducing, living cell. Before that landmark can be reached, several challenges must be met.
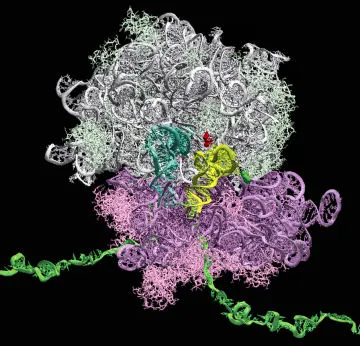
Most significant, perhaps, is the difficulty of making ribosomes. While many enzymes are involved in protein synthesis, ribosomes are the cell’s main protein factories. Each ribosome is a complex assembly of proteins and RNA molecules, which are similar to DNA but with only one strand instead of a double helix. Scientists have made bacterial ribosomes in the lab, but only at temperatures too high for liposomes to endure.
Presumably, since natural cells can make ribosomes, scientists could simply replicate those cells’ natural machinery in their synthetic liposomes, and ribosomes would spring forth. But ribosome creation requires many RNA-modification enzymes that are still unknown.
In the beginning
Conjuring simple life from nonlife in a dish would no doubt raise comparisons with the emergence of the first living cells on Earth about 4 billion years ago.
“The closer we get to a simple system, the closer we get to a good early cell model,” Murtas said at Synthetic Biology 3.0.
But the analogy is not perfect. As Murtas points out, “Using [modern] biological molecules, I find it hard to believe that an early simple cell … can ever have existed with only 30 to 40 genes.” But how could it be possible for the first cells to have already evolved more than 40 working genes?
The synthetic minimal cell envisioned by Forster and Church wouldn’t reveal much about how life began. For one thing, the protein and gene “parts” from which scientists would build such a cell are all modern molecules shaped by millions or billions of years of evolution. Assembling these parts into a simpler system would be like building a Model T using Honda parts: It wouldn’t turn back the hands of time on the parts themselves.
More fundamentally, though, the earliest living cells on Earth didn’t have DNA or proteins at all, most scientists think. Instead, those cells most likely relied on simpler RNA molecules to act as both catalysts for chemical reactions (as proteins do) and carriers of genetic information (like DNA) (SN: 4/7/01, p. 212).
“We’re all pretty confident that there was an early phase in the origin of life in which cell functions were carried out by RNA,” says Jack Szostak of Harvard Medical School in Boston. The inner workings of such a cell would be radically different from those of any cell made using modern parts.
Szostak and others are pursuing a third way to create a human-made cell that would more closely resemble the origin of life. In its idealized form, such a cell would consist of a simple lipid membrane housing a single type of RNA-like molecule that is capable of self-replication.
The actual design might end up being just that simple.
Research published by Szostak and his colleagues in 2004 showed that an internal pressure on the membrane caused by a growing number of RNA molecules is enough to cause the membrane to take up lipid molecules from the environment and enlarge.
“You can do this with just physical principles,” Szostak says.
The challenge, then, is to design an RNA or similar molecule that can self-replicate efficiently. Trying to do so has already yielded some lessons about the origin of life.
The earliest self-replicating molecules needed to overcome a few hurdles that also vex researchers. RNA degrades relatively quickly, so forming RNAs that are long enough to carry much genetic information is difficult. And during self-replication, a newly formed RNA strand becomes bound to the original strand like the two sides of a zipper. Scientists are still searching for efficient ways to pull off the new strand so that the original template can repeat the process.
Because of these difficulties, Szostak and others are experimenting with a variety of novel RNA-like molecules. Rather than testing every possible variant by hand, scientists such as Gerald F. Joyce of the Scripps Research Institute in La Jolla, Calif., set up experimental conditions that guide the rapid evolution of these molecules toward the desired traits. “You select for molecules that do what you want, then you amplify the ones that do that, then you repeat the cycle,” Szostak explains.
This technique has already produced RNA-like molecules that can reproduce, but the process is still inefficient and error prone. It’s hard to predict how long it will take for scientists to stumble upon a viable candidate, but Szostak thinks it may not be far off. “The magnitude of the improvement that’s needed is not huge,” he says.
If they succeed, scientists might be able to observe how this primitive cell evolves more-complex traits over time, like watching a reenactment of the origin of life.
For all three of these approaches to making synthetic or semisynthetic cells, it seems to be a question not of if, but when. Once people learn how to make living cells from scratch in the lab, perhaps life there will routinely emerge out of nonlife, though not quite in the way that Aristotle imagined.