Computers employ a variety of schemes to check whether a chunk of digital information–transmitted as a message, stored in a database, or functioning as a set of instructions–remains error-free. Such error-detection codes would detect, for example, the change of one bit from 0 to 1 or 1 to 0 in corrupted data.
These codes typically rely on the addition of one or more bits to a data string to carry the error-detection information. In a simple binary parity check, a single additional bit would represent whether the total number of “1s” in a given data string is even (1) or odd (0). For example, in the string 1001 0100, the total number of “1s” is odd, so the appended bit would be 1, to generate the new string 1001 0100 1. If one bit changes, the number of “1s” would no longer be odd, and the parity bit would be incorrect, allowing detection of the error.
Strands of DNA also carry information–of the genetic sort–encoded in their chemical structure. Chemist Dónall A. Mac Dónaill of Trinity College in Dublin, Ireland, has now shown that patterns inherent in the chemical makeup of DNA correspond to a digital error-detecting code. His report appears in the Sept. 12 Chemical Communications.
A single strand of DNA consists of a chain of simpler molecules called bases (or nucleotides), which protrude from a sugar-phosphate backbone. The four varieties of bases are known as adenine (A), thymine (T), guanine (G), and cytosine (C). These bases constitute the nucleotide alphabet.
Why are there precisely four nucleotides, and why are these particular nucleotides the critical ones? “Not all combinations of nucleotides would be equally error-resistant, allowing selection pressure to select an optimal alphabet with respect to informatics,” Mac Dónaill suggests.
Any strand of DNA will adhere tightly to its complementary strand, in which T substitutes for A, G for C, and vice versa. For example, a single-stranded DNA segment consisting of the base sequence TAGCC will stick to a section of another strand made up of the complementary sequence ATCGG. The links (hydrogen bonds) between pairs of bases are responsible for binding together two strands to form the characteristic double helix of a DNA molecule. The same linking mechanism comes into play when genetic information stored in DNA is read out in living cells to produce protein molecules and to replicate that information when cells divide.
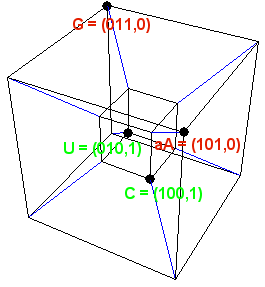
The hydrogen bonding that occurs between nucleotides is highly selective. These bonds are characterized by particular patterns of chemical groups known as donors (hydrogen atoms) and acceptors (lone pairs of electrons). Mac Dónaill represented the donor-acceptor pattern of each nucleotide as a string of three bits. For example, if a donor is (arbitrarily) represented as 1 and an acceptor as 0, the pattern 100 would encode C and 011 would encode G.
At the same time, A and G belong to the purine group of molecules; T and C belong to the pyrimidine group. If a purine is represented by 0 and a pyrimidine by 1, the full code for C would be 100,1 and for G would be 011,0.
In all, there are 16 possible four-bit numbers, and the nucleotides that go into DNA represent only four of these combinations. Moreover, it turns out that each nucleotide has an even number of “1s.” In informatics parlance, each nucleotide would be a codeword, and all the codewords have the same parity.
“A code in which all codewords have the same parity is termed a parity code, and possesses simple but effective error-resistant properties,” Mac Dónaill remarks. In this case, there’s no appended parity bit, but because the codewords all have the same parity, the code can still serve an error-detection function.
“Thus, it would appear that in nature the purine/pyrimidine nature of a nucleotide is strictly and intriguingly related to the [donor/acceptor] pattern as a parity bit,” Mac Dónaill says.
Was this accidental or shaped by selection through evolutionary advantage? Mac Dónaill favors the latter possibility. “Factors other than physicochemical issues alone shaped the natural nucleotide alphabet,” he suggests.
Indeed, nature may very well have anticipated modern error-coding theory to help ensure that the right things happen in the right way.