No gene is an island
Even as biologists catalog the discrete parts of life forms, an emerging picture reveals that life’s functions arise from interconnectedness
The gene p53 has long been singled out as an anticancer hero. It is a critical tumor fighter. A person or lab animal develops a tumor much faster without the gene than with it.
But p53 could be dangerous if left to act alone.
What really gives the gene its power is its network within a cell. Cells must guard against the constant threat of becoming cancerous, a change sometimes triggered by damage to the cells’ DNA. The p53 gene is embedded in a network of interacting genes and proteins, and this web of interactions provides sophisticated control of p53, keeping it in check. Such control is important because p53 is a double-edged sword: It can either promote DNA repair or—if the DNA damage is too severe—trigger the cancer-prone cell to self-destruct (thus sparing the rest of the body from getting cancer).
Balancing these two functions is critical for keeping cancer at bay.
Single genes get the limelight, but a gene or protein rarely acts alone. Now that biology’s centuries-long quest to dissect living things into ever-smaller parts is reaching its logical conclusion—the comprehensive list of every gene, protein and molecule in cells—some scientists are beginning the daunting task of putting Humpty Dumpty back together again. Little by little, research is revealing how all these parts are connected in vast networks, networks in which genes and proteins interact in living cells to produce cell behaviors.
And a cell’s behavior is crucial to its survival. Whether a single cell floating in the sea or a cell within a plant or person, a cell must do the right thing at the right time in the right way or it might simply die. Whatever the cell’s fancy internal mechanisms may be, the end result—the cell’s behavior—is all that matters.
“The community has produced a lot of information on the parts list, with sequenced genomes, proteomics, metabolics,” says Ilya Shmulevich, a systems biologist at the Institute for Systems Biology in Seattle. “There’s so much data that is produced, and right now what needs to happen is all of this needs to come together.”
By looking at how all the parts of a cell are networked, researchers are finding subnetworks that behave much like basic components of electronic circuits, such as switches, blinkers and buzzers.
Other recent work suggests that entire cell networks balance themselves at the threshold separating order from chaos.
When applied to an organism’s inner workings, a network perspective can solve problems that a gene-centric approach can’t—such as understanding the growth of a sea urchin embryo or how the segmentation of insect bodies is encoded.
Many scientists also think that understanding human cells at the network level is essential for developing drugs to treat complex diseases such as diabetes and cancer.
“These harder diseases remain unsolved because they aren’t caused by individual proteins. They’re diseases of the network,” said systems biologist Hans Westerhoff of the University of Manchester in England, in August during the International Conference on Systems Biology in Gothenburg, Sweden.
Using network analysis, Westerhoff’s team found that the best target in a cell for anticancer drugs was not the one researchers suspected. So with the cellular parts lists from genomics in hand, scientists are beginning to connect the dots.
Mapping motifs
Networks are full of surprises.
“They often do things you didn’t think they would do, and often they refuse to do things that you thought they would do,” says John Tyson, a computational biologist at Virginia Tech in Blacksburg. That’s because networks are more than the sum of their parts. A network’s architecture—the pattern of interactions among its parts—strongly influences its overall behavior in ways that can’t be predicted based on the parts alone. And some connection patterns give rise to nonintuitive, nonlinear behaviors.
In a diagram of a gene and protein network, each dot represents a certain gene or protein, and a line between dots means that those two parts interact in some way. Perhaps one gene activates another, or a protein binds to and inhibits another protein. A basic network diagram looks a bit like a game of connect the dots that ends in a jumbled mess instead of a pretty picture.
It’s a simple premise, but once you start wiring together a few genes or proteins this way, the behavior of the resulting network proves to be quite sophisticated. The right “motif,” or pattern of connections among a few proteins, will behave like a switch, buzzer, sniffer or blinker.
“You can understand complicated protein interaction networks in terms of these little motifs that are hooked together, like you can construct an electronic circuit,” Tyson says. “There’s been a lot of progress here recently.”
Combining even a few such motifs in a small subnetwork can generate useful behaviors. For example, the tumor-suppressor gene p53 is controlled by just a few motifs that combine to make p53 behave much like a ticking time bomb.
Genes carry the codes for specific proteins, and the amount of a protein that a gene makes at any time is defined as the gene’s activity. After DNA damage occurs in a cell, p53 springs to life, and its activity—and thus the concentration of the p53 protein it encodes—begins to oscillate slowly from more to less and back again, like a throbbing warning light. The number of oscillations depends on how bad the damage is. If the DNA hasn’t been fixed by the time the oscillations stop, p53 activity begins to ramp up slowly and steadily until it reaches a point that triggers the cell’s suicide machinery.
In a network diagram, p53 is connected with two other genes, MDM2 and ATM, in a motif that could be called a damper: a negative feedback loop. The classic example of this kind of feedback loop is a thermostat. If temperatures get too high, the air conditioning switches on and cools things down, and when things get too cool, the air conditioning switches off to let the room warm back up. So the temperature oscillates around some desired point. For genes in a network, a similar motif occurs when one gene activates another gene, which in turns inhibits the first gene.
Negative feedback loops with MDM2 and ATM can explain p53’s oscillation behavior after DNA damage, Galit Lahav of the systems biology department at HarvardMedicalSchool in Boston and his colleagues reported in the May 9 issue of Molecular Cell.
As time runs out, a second part of the network—a positive feedback loop between p53 and the genes PTEN, PIP3 and Akt—overcomes this first motif. Positive feedback loops could be called amplifiers: They cause an increase to keep increasing. Genes in this second motif cut off the damper by blocking the connection between MDM2 and p53, Tomasz Lipniacki of the Institute of Fundamental Technological Research in Warsaw, Poland, and his colleagues reported in the Sept. 21 Journal of Theoretical Biology. This change switches off the thermostat, so the positive feedback loop can drive up p53 activity until it triggers cell suicide.
Zoom out
Somewhat larger networks containing a dozen or more motifs can control even more complex processes, such as that by which insect embryos develop body segments. Different species develop their segmented body plans in different ways, but the genes involved in this segmentation are roughly the same across most insect species. Koichi Fujimoto and his colleagues at the University of Tokyo wondered whether three different development plans could be coded not in the genes themselves, but in the way the genes are connected in the network.
Fujimoto’s team used a computer to “evolve” hundreds of simulated gene networks, mutating each network repeatedly until it produced one of the three development plans. Looking at the resulting networks, the researchers found that each of the development plans corresponded to one of three network characteristics: Many feed-forward loops, at least one negative feedback loop or interwoven loops of both types.
While the work was based on networks simulated in a computer, comparison with the known gene networks of the fruit fly and the red flour beetle showed that those insects had the same network patterns predicted by the simulations, Fujimoto’s team reported online July 23 in PLoS ONE.
By understanding in detail how networks give rise to cell behavior in health, “we can also understand how they fail in disease,” Shmulevich says. “The future is that we’re going to be looking in the blood for various organ-specific signatures of disease so that we can detect how the networks have been perturbed.”
Developing drugs is currently a trial and error game, he says: “Having systems-level understanding of biological processes and being able to predict how they’ll respond to various changes will really help drug development. We can drive the system toward a certain state or away from a certain state.”
That’s still a far-off goal, but some research on still-larger networks suggests that this kind of control is possible.
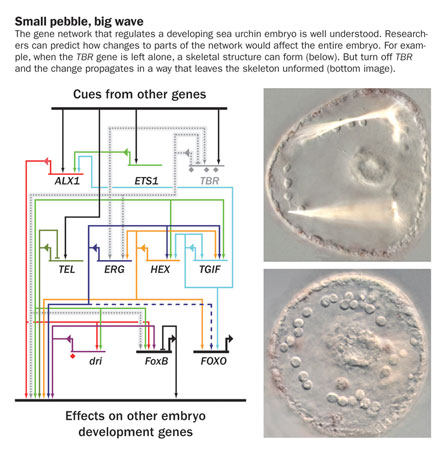
The entire map
Perhaps the best-understood large gene network is the one controlling the development of sea urchin embryos. Over the past four decades, Eric Davidson, a cell biologist at California Institute of Technology in Pasadena, has been studying sea urchin development. In the past few years he’s assembled a detailed model of the gene networks that orchestrate the entire process of intestine formation. In the April 22 Proceedings of the National Academy of Sciences, Davidson and his colleagues also published a detailed description of the networks that steer some of the urchin’s cells into becoming skeleton-forming cells.
These mathematical models chart the networks that regulate the activity of the sea urchin’s genes. The models are complete enough to accurately predict how a change to the network will affect the embryo’s development. If such thorough network models can eventually be made for human cells, scientists may be able to skillfully alter the cells’ networks to treat diseases such as cancer, diabetes and autoimmune disorders.
“The complex diseases are the ones that we have to understand on a systems level,” Shmulevich says.
Zooming the picture out farther provides a view of how the network behaves as a whole.
For example, research reported in August at the systems biology conference in Sweden suggests that gene networks obey a principle called the “law of conservation of fragility.” If disturbing one gene in a network has a large effect on the network’s overall functioning, that gene is considered fragile. If the effect on the network is small or negligible, the gene is not fragile. The law says that if you add up the fragility values for all the genes or proteins in a network, the total will always be the same, according to a study presented by Manchester’s Westerhoff.
So if a change in the network makes some genes less fragile, the law tells you that there must be at least one gene that becomes more fragile to compensate—conservation of fragility (“Finding health in fragility,” SN Online: 8/25/08).
“The system may be robust in some places, but it must also be fragile in some other places,” Westerhoff said at the meeting. “If you’re developing a drug, you might want to target these fragile places.”
This principle suggests a surprising target for anticancer drugs. A gene called RAF is overactive in many kinds of cancer, and so might stand out as a potential target. But Westerhoff and his colleagues showed that this gene is actually more fragile in healthy cells than cancer cells. So a drug targeting that gene would damage healthy cells more than cancerous ones. Because of conservation of fragility, some other gene must be more fragile in the cancer cells and thus would make a more effective target for a drug, Westerhoff says.
For very large networks encompassing entire cells, scientists are also learning how to watch the whole network’s behavior over time.
The basic idea is to represent the condition of the complete network at one moment in time as a point in space, called “state space.” If the network had only three genes, the state space could be represented as a 3-D mathematical graph. The point would have three coordinates that represent the activity levels of the three genes—that is, how actively those genes churn out copies of proteins. For a network with hundreds or thousands of genes, the space would have hundreds or thousands of dimensions. Such high-dimensional space can’t be visualized but can be dealt with mathematically.
As long as the activity of these genes—and hence the cell’s behavior—remains the same, the point will hold still. If some genes become more or less active over time, causing a change in the behavior of the cell, the point will move in state space to reflect that change.
Mathematicians have shown that complex networks with many feedback loops—like the networks in cells—can behave in one of a few ways in this state space over time. Some network architectures will gravitate toward a stable point and stay there. The cell’s behavior becomes unchanging. Other networks will home in on a repeating cycle, such as a circle in state space, and then follow that pattern endlessly. For this kind of cell, steady oscillation would be its most stable state. Scientists call both states ordered behavior.
But tweak the architecture of an ordered network just a little bit and the system can cross over into chaotic behavior. In this case, “chaos” doesn’t mean mayhem or randomness. It just means that the network will follow a meandering path through state space that, unlike ordered behavior, never repeats. That path isn’t truly random, but it is unpredictable because even the tiniest, immeasurable difference in its state at one moment can gradually swell until it alters the course of the entire network—like the famous butterfly that causes a hurricane by flapping its wings.
It turns out that ordered and chaotic behavior are both dangerous for a cell. Ordered behavior is too stable, so the cell can’t respond to challenges in its environment. Chaotic behavior balloons disturbances until they overrun the cell, preventing the cell from keeping its interior suitable for the chemistry of life.
But there’s a Goldilocks zone at the threshold between order and chaos called criticality.
“It’s been hypothesized for a long time that living systems are critical, and what we’ve done is show for the first time on a molecular level that that’s true for these cells,” Shmulevich says.
Shmulevich and his colleagues showed that a kind of immune system cell called a macrophage does indeed operate in the Goldilocks zone between order and chaos, thus striking the right balance between stability and flexibility. The scientists stimulated molecules on the cell’s surface that detect foreign bacteria. By watching how this stimulation spread through the cell and affected the activity levels of all the cell’s genes, Shmulevich’s team found that the change in gene activity neither petered out immediately nor amplified over time. This pattern is exactly what is expected for a cell at criticality, the researchers reported in February in PNAS.
“The perturbation can propagate and can travel through the system, but it’s not going to swamp the system and make it useless,” Shmulevich says.
His group later repeated this experiment on cells from five species representing four of the kingdoms of life: bacteria, yeast, plants and animals. Once again, cells from each of the kingdoms operated at criticality, the team reported online June 18 in PLoS ONE.
This work begins to reveal the hidden dynamics that emerge when genes and proteins act together in networks, says Leroy Hood, president of Seattle’s Institute for Systems Biology. But Hood adds that, even after the mammoth task of thoroughly mapping the networks in a human cell is done, still to be understood is how that cell’s networks integrate into a larger, different kind of network: the billions of interacting cells in an organ such as the heart or—shudder—the brain.
“Boy are we at the very, very beginning of that,” Hood says.