Cell’s core pore structure solved
From Washington, D.C., at a meeting of the American Society for Cell Biology
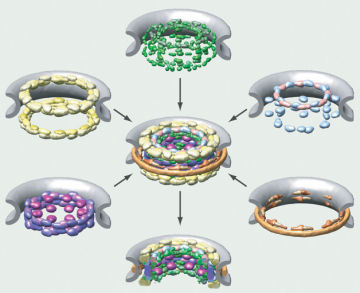
In a challenge that’s like solving a three-dimensional jigsaw puzzle, scientists have worked out how 456 proteins fit together to form a doughnut-shaped gateway to the cell’s nucleus.
Many such gateways cover the nucleus like polka dots, and they control what protein can pass through to reach the DNA inside. Knowing the structure of the gateway—called a nuclear-pore complex (NPC)—could improve scientists’ understanding of why some large molecules can enter the nucleus and others cannot.
“[The NPC is] one of the biggest stable protein complexes in the cell,” says Frank Alber of the University of California in San Francisco (UCSF). Even finding the structure of pores in a cell’s outer membrane can be challenging, and those pores typically consist of less than a dozen proteins—far fewer than the NPC.
Solving the puzzle required pulling together many different kinds of data about the proteins. “None of the types of data on their own can tell us the structure” Alber says. “It’s a bit like a crossword puzzle” in which each clue is ambiguous, but the overlapping answers restrict what the other answers can be, thus narrowing the possibilities to a single solution.
Alber and his colleagues at UCSF and Rockefeller University in New York used different kinds of data to place constraints on where each protein could be located relative to other proteins. For example, the researchers repeatedly fragmented the complex into small pieces, and then observed which clumps of proteins remained tightly bound to identify adjacent proteins. And electron microscope images of an entire pore revealed its overall shape. Altogether, the scientists amassed about 10,000 constraints on the proteins’ locations.
Starting from random swarms of proteins, the scientists then used computers to gradually find stable protein arrangements that met all of the constraints. Repeating this process thousands of times produced the most likely position for each protein.
The researchers studied NPCs from yeast cells, which have nuclei, just as human, animal, plant, and algae cells do. In principle, the new computational method could be applied to the human NPC, Alber says, although it is considerably larger than the yeast version.