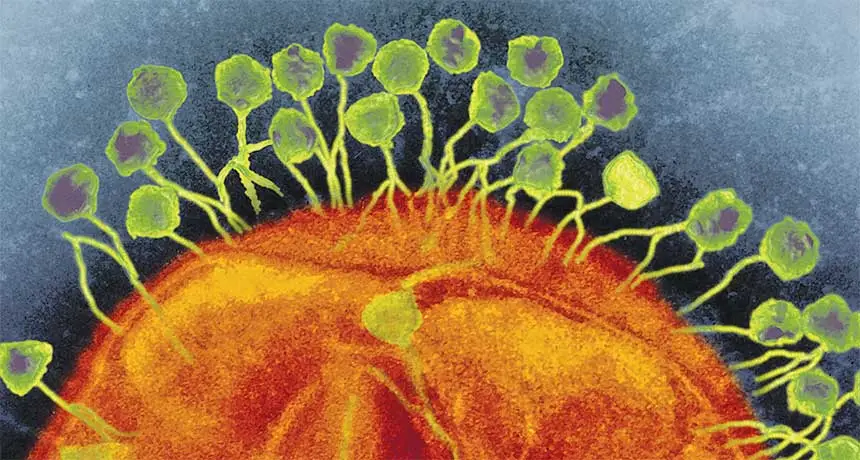
WEAPONS OF MASS EVOLUTION Bacteria and archaea armed with CRISPR systems have been at war with viruses for eons. Here, hordes of viruses known as phages assault a bacterium to turn it into a virus-making factory.
AMI IMAGES/SCIENCE SOURCE
It is the dazzling star of the biotech world: a powerful new tool that can deftly and precisely alter the structure of DNA. It promises cures for diseases, sturdier crops, malaria-resistant mosquitoes and more. Frenzy over the technique — known as CRISPR/Cas9 — is in full swing. Every week, new CRISPR findings are unfurled in scientific journals. In the courts, universities fight over patents. The media report on the breakthroughs as well as the ethics of this game changer almost daily.
But there is a less sequins-and-glitter side to CRISPR that’s just as alluring to anyone thirsty to understand the natural world. The biology behind CRISPR technology comes from a battle that has been raging for eons, out of sight and yet all around us (and on us, and in us).
The CRISPR editing tool has its origins in microbes — bacteria and archaea that live in obscene numbers everywhere from undersea vents to the snot in the human nose. For billions of years, these single-celled organisms have been at odds with the viruses — known as phages — that attack them, invaders so plentiful that a single drop of seawater can hold 10 million. And natural CRISPR systems (there are many) play a big part in this tussle. They act as gatekeepers, essentially cataloging viruses that get into cells. If a virus shows up again, the cell — and its offspring — can recognize and destroy it. Studying this system will teach biologists much about ecology, disease and the overall workings of life on Earth.
But moving from the simple, textbook story into real life is messy. In the few years since the defensive function of CRISPR systems was first appreciated, microbiologists have busied themselves collecting samples, conducting experiments and crunching reams of DNA data to try to understand what the systems do. From that has come much elegant physiology, a mass of complexity, surprises aplenty — and more than a little mystery.
Spoiled yogurt
The biology is complicated, and its basic nuts and bolts took some figuring out. There are two parts to CRISPR/Cas systems: the CRISPR bit and the Cas bit. The CRISPR bit — or “clustered regularly interspaced short palindromic repeats” — was stumbled on in the late 1980s and 1990s. Scientists then slowly pieced the story together by studying microbes that thrive in animals’ guts and in salt marshes, that cause the plague and that are used to make delicious yogurt and cheese.
None of the scientists knew what they were dealing with at first. They saw stretches of DNA with a characteristic pattern: short lengths of repeated sequence separated by other DNA sequences now known as spacers. Each spacer was unique. Because the roster of spacers could differ from one cell to the next in a given microbe species, an early realization was that these differences could be useful for forensic “typing” — investigators could tell whether food poisoning cases were linked, or if someone had stolen a company’s yogurt starter culture.
But curious findings piled up. Some of those spacers, it turned out, matched the DNA of phages. In a flurry of reports in 2005, scientists showed, to name one example, that strains of the lactic acid bacterium Streptococcus thermophilus contained spacers that matched genetic material of phages known to infect Streptococcus. And the more spacers a strain had, the more resistant it was to attack by phages.
This began to look a lot like learned or adaptive immunity, akin to our own antibody system: After exposure to a specific threat, your immune system remembers and you are thereafter resistant to that threat. In a classic experiment published in Science in 2007, researchers at the food company Danisco showed it was so. They could see new spacers added when a phage infected a culture of S. thermophilus. Afterward, the bacterium was immune to the phage. They could artificially engineer a phage spacer into the CRISPR DNA and see resistance emerge; when they took the spacer away, immunity was lost.
This was handy intel for an industry that could find whole vats of yogurt-making bacteria wiped out by phage infestations. It was an exciting time scientifically and commercially, says Rodolphe Barrangou of North Carolina State University in Raleigh, who did a lot of the Danisco work. “It was not just discovering a cool system, but also uncovering a powerful phage-resistance technology for the dairy industry,” he says.
The second part of the CRISPR/Cas system is the Cas bit: a set of genes located near the cluster of CRISPR spacers. The DNA sequences of these genes strongly suggested that they carried instructions for proteins that interact with DNA or RNA in some fashion — sticking to it, cutting it, copying it, unraveling it. When researchers inactivated one Cas gene or another, they saw immunity falter. Clearly, the two bits of the system — CRISPR and Cas — were a team.
It took many more experiments to get to today’s basic model of how CRISPR/Cas systems fight phages — and not just phages. Other types of foreign DNA can get into microbes, including circular rings called plasmids that shuttle from cell to cell and DNA pieces called transposable elements, which jump around within genomes. CRISPRs can fend off these intruders, as well as keep a microbe’s genome in tidy order.
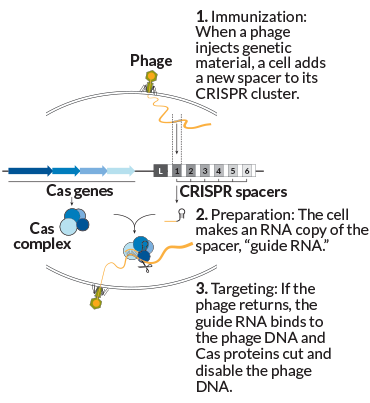
The process works like this: A virus injects its genetic material into the cell. Sensing this danger, the cell selects a little strip of that genetic material and adds it to the spacers in the CRISPR cluster. This step, known as immunization or adaptation, creates a list of encounters a cell has had with viruses, plasmids or other foreign bits of DNA over time — neatly lined up in reverse chronological order, newest to oldest.
Older spacers eventually get shed, but a CRISPR cluster can grow to be long — the record holder to date is 587 spacers in Haliangium ochraceum, a salt-loving microbe isolated from a piece of seaweed. “It’s like looking at the last 600 shots you had in your arm,” says Barrangou. “Think about that.”
New spacer in place, the microbe is now immunized. Later comes targeting. If that same phage enters the cell again, it’s recognized. The cell has made RNA copies of the relevant spacer, which bind to the matching spot on the genome of the invading phage. That “guide RNA” leads Cas proteins to target and snip the phage DNA, defanging the intruder.
Researchers now know there are a confetti-storm of different CRISPR systems, and the list continues to grow. Some are simple — such as the CRISPR/Cas9 system that’s been adapted for gene editing in more complex creatures (SN: 4/15/17, p. 16) — and some are elaborate, with many protein workhorses deployed to get the job done.
Those who are sleuthing the evolution of CRISPR systems are deciphering a complex story. The part of the CRISPR toolbox involved in immunity (adding spacers after phages inject their genetic material) seems to have originated from a specific type of transposable element called a casposon. But the part responsible for targeting has multiple origins — in some cases, it’s another type of transposable element. In others, it’s a mystery.
The downsides
Given the power of CRISPR systems to ward off foes, one might think every respectable microbe out there in the soils, vents, lakes, guts and nostrils of this planet would have one. Not so.
Numbers are far from certain, partly because science hasn’t come close to identifying all the world’s microbes, let alone probe them all for CRISPRs. But the scads of microbial genetic data accrued so far throw up interesting trends.
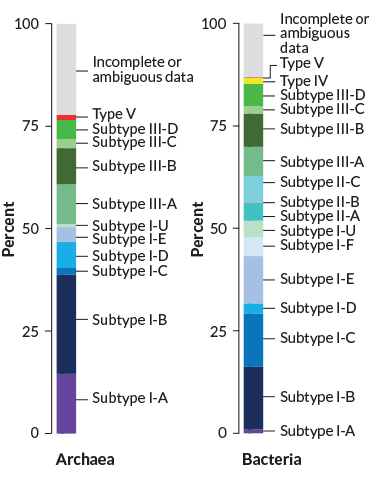
Tallies suggest that CRISPR systems are far more prevalent in known archaea than in known bacteria — such systems exist in roughly 90 percent of archaea and about 35 percent of bacteria, says Eugene Koonin, a computational evolutionary biologist at the National Institutes of Health in Bethesda, Md. Archaea and bacteria, though both small and single-celled, are on opposite sides of the tree of life.
Perhaps more significantly, Koonin says, almost all the known microbes that live in superhot environments have CRISPRs. His group’s math models suggest that CRISPR systems are most useful when microbes encounter a big enough variety of viruses to make adaptive memory worth having. But if there’s too much variety, and viruses are changing very fast, CRISPRs don’t really help — because you’d never see the same virus again. The superhot ecosystems, he says, seem to have a stable amount of phage diversity that’s not too high or low.
And CRISPR systems have downsides. Just as people can develop autoimmune reactions against their own bodies, bacteria and archaea can accidentally make CRISPR spacers from bits of their own DNA — and risk chewing up their own genetic material. Researchers have seen this happen. “No immunity comes without a cost,” says Rotem Sorek, a microbial genomicist at the Weizmann Institute of Science in Rehovot, Israel.
But mistakes are rare, and Sorek and his colleagues recently figured out why in the microbe they study. The researchers reported in Nature in 2015 that CRISPR spacers are created from linear bits of DNA — and phage DNA is linear when it enters cells. The bacterial chromosome is protected because of its circular form. Should it break and become linear for a spell, such as when it’s being replicated, it contains signals that ward off the Cas proteins.
There are other negatives to CRISPR systems. It’s not always a bonus to keep out phages and other invaders, which can sometimes bring in useful things. Escherichia coli O157:H7, of food poisoning fame, can make humans sick because of toxin genes it harbors that were brought in by a phage, to name just one of myriad examples. Even CRISPR systems themselves are spread around the microbial kingdom via phages, plasmids or transposable elements.
For microbes that lack CRISPR systems, there are many other ways to repel foreign DNA — as much as 10 percent of a microbial genome may be devoted to hawkish warfare, and new defense systems are still being uncovered.
Countermeasures
The war between bacteria and phages is two-sided, of course. Just as a microbe wants to keep doors shut to protect its genetic integrity and escape destruction, the phage wants in.
And so the phage fights back against CRISPRs. It genetically morphs into forms that CRISPRs no longer recognize. Or it designs bespoke artillery. Microbiologist Joe Bondy-Denomy, now at the University of California, San Francisco, happened upon such customized weapons as a grad student in the lab of molecular microbiologist Alan Davidson at the University of Toronto. The team knew that the bacterium Pseudomonas aeruginosa, which lives in soil and water and can cause dangerous infections, has a vigorous CRISPR system. Yet some phages didn’t seem fazed by it.
That’s because those phages have small proteins that will bind to and interfere with this or that part of the CRISPR machinery, such as the Cas enzyme that cuts phage DNA. The binding disables the CRISPR system, the researchers reported in 2015 in Nature. Bondy-Denomy and others have since found anti-CRISPR genes in other phages and other kinds of interloping DNA. The genes are so common, Davidson says, that he wonders how many CRISPR systems are truly active.
In an especially bizarre twist, microbiologist Kimberley Seed of the University of California, Berkeley found a phage that carries its own CRISPR system and uses it to fight back against the cholera bacterium it invades, she and colleagues reported in 2013 in Nature. It chops up a segment of bacterial DNA that normally inhibits phage infection.
Of course, in this never-ending scuffle one would expect the microbes to again fight back against the phages. “It’s something I often get asked: ‘Great, the anti-CRISPRs are there, so where are the anti-anti-CRISPRs?’ ” Bondy-Denomy says. Nobody has found such things yet.
Evolution drivers
It’s one thing to study CRISPR systems in well-controlled lab settings, or in just one type of microbe. It’s another to understand what all the various CRISPRs do to shape the ecosystem of a bubbling hot spring, human gut, diseased lung or cholera-tainted river. Estimates of CRISPR abundance could drop as more sampling is done, especially of dark horse microbes that researchers know little about.
In a 2016 report in Nature Communications, for example, geomicrobiologist Jill Banfield of UC Berkeley and colleagues detected 1,724 microbes in Colorado groundwater that had been treated to boost the abundance of types that are difficult to isolate. CRISPR systems were much rarer in this sample than in databases of better-known microbes.
Tallying CRISPRs is just the start, of course. Microbial communities — including those inside our own guts, where there are plenty of CRISPR systems and phages — are dynamic, not frozen. How do CRISPRs shape the evolution of phages and microbes in the wild? Banfield’s and Barrangou’s labs teamed up to watch as S. thermophilus and phages incubated together in a milk medium for hundreds of days. The team saw bacterial numbers fall as phages invaded; then bacteria acquired spacers against the phage and rallied — and phage numbers fell downward in turn. Then new phage populations sprang up, immune to S. thermophilus defenses because of genetic changes. In this way, the researchers reported in 2016 in mBio, CRISPRs are “one of the fundamental drivers of phage evolution.”
CRISPR systems can be picked up, dropped, then picked up again by bacteria and archaea over time, perhaps as conditions and needs change. The bacterium Vibrio cholerae is an example of this dynamism, as Seed and colleagues reported in 2015 in the Journal of Bacteriology. The older, classical strains of this medical blight harbored CRISPRs, but these strains went largely extinct in the wild in the 1960s. Strains that cause cholera today do not have CRISPRs.
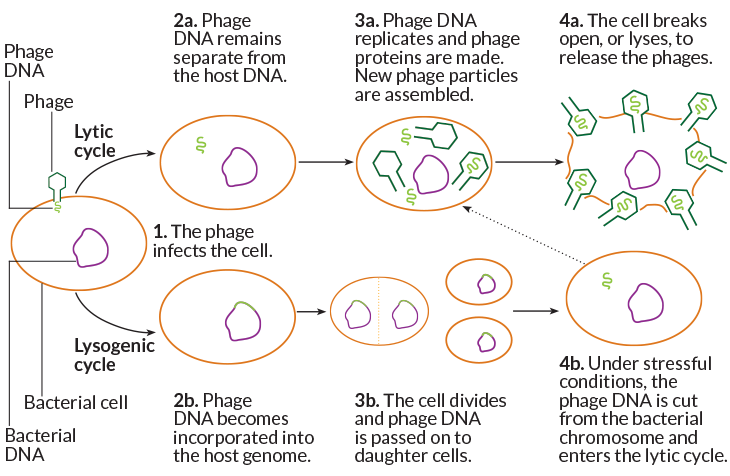
Nobody knows why, Seed says. But scientists stress that it is a mischaracterization to paint the relationship between microbes and phages, plasmids and transposable elements as a simplistic war. Phages don’t always wreak havoc; they can slip their genomes quietly into the bacterial chromosome and coexist benignly, getting copied along with the host DNA. Phages, plasmids and transposable elements can confer new, useful traits — sometimes even essential ones. Indeed, such movement of DNA across species and strains is at the heart of how bacteria and archaea evolve.
So it’s about finding balance. “If you incorporate too much foreign DNA, you cannot maintain a species,” says Luciano Marraffini, a molecular microbiologist at the Rockefeller University in New York City whose work first showed that DNA-cutting was key to CRISPR systems. But you do need to let some DNA in, and it’s likely that some CRISPR systems permit this: The system he studies in Staphylococcus epidermidis, for example, only goes after phages that are in their cell-killing, or lytic, state, he and colleagues reported in 2014 in Nature.
Story continues after graphic
Beyond defense
One thing is very clear about CRISPR systems: They are perplexing in many ways. For a start, the spacers in a microbe should reflect its own, individual story of the phages it has encountered. So you’d think there would be local pedigrees, that a bacterium sampled in France would have a different spacer cluster from a bacterium sampled in Argentina. This is not what researchers always see.
Take the nasty P. aeruginosa. Rachel Whitaker, a microbial population biologist at the University of Illinois at Urbana-Champaign, studies Pseudomonas samples collected from people with cystic fibrosis, whose lungs develop chronic infections. She’s found no sign that two patients living close to each other carry more-similar P. aeruginosa CRISPRs than two patients thousands of miles apart. Yet surely one would expect nearby CRISPRs to be closer matches, because the Pseudomonas would have encountered similar phages. “It’s very weird,” Whitaker says.
Others have seen the same thing in heat-loving bacteria sampled from very distant bubbling hot springs. It’s as if scientists don’t truly understand how bacteria spread around the world — there could be a strong effect of far-flung passage by air or wind, says Konstantin Severinov, who studies CRISPR systems at Rutgers University in New Brunswick, N.J.
Another weirdness is the differing vigor of CRISPR systems. Some are very active. Molecular biologist Devaki Bhaya of the Carnegie Institution for Science’s plant biology department at Stanford University sees clear signs that spacers are frequently added and dropped in the cyanobacteria of Yellowstone’s hot springs, for example. But other systems are sluggish, and E. coli, that classic workhorse of genetics research, has a respectable-looking CRISPR system — that is switched off.
It may have been off for a long time. Some 42,000 years ago, a baby woolly mammoth died in what is now northwestern Siberia. The remains, found in 2007, were so well-preserved that the intestines were intact and E. coli DNA could be extracted.
In research published in Molecular Ecology in January, Severinov’s team found surprising similarities between the spacers in the mammoth-derived E. coli CRISPR cluster and those in modern-day E. coli. “There was no turnover in all that time,” Severinov marvels. If the CRISPR system isn’t active, why does E. coli bother to keep it?
That quandary leads neatly to what some researchers refer to as an intellectually “scandalous situation.”
In some cases, the genetic sequence of spacers nicely matches phage DNA. But overall, only a fraction (around 1 to 2 percent) of the spacers scientists know about have been matched to a virus or a plasmid. In E. coli, the spacers don’t match common, classic phages known to infect the bacterium. “Is it the case that there is a huge, unknown amount of viral dark matter in the world?” says Koonin — or are phages evolving superfast? “Or is it something completely different?”
Faced with this conundrum, some researchers strongly suspect — and have evidence — that CRISPR systems may do more than defend; they may have other jobs. Communication, perhaps. Or turning genes on and off.
But some microbes’ CRISPR sequences do make sense, especially if looking at the spacers most recently added, and others may be clues to phages still undiscovered. So even as they scratch their heads about many things CRISPR, scientists are also excited by the stories CRISPR clusters can tell about the viruses and other bits of DNA that bacteria and archaea encounter and that they choose, for whatever reason, to note for the record. What do microbes pay attention to? What do they ignore?
CRISPRs offer a bright new window on such questions and, indeed, already are unearthing novel phages and facts about who infects whom in the microscopic world.
“We can catalog everything that’s out there. But we don’t really know what matters,” says Bondy-Denomy. “CRISPRs can help us understand.”
Rosie Mestel is a freelance writer based in Los Angeles.
This article appears in the April 15, 2017, issue of Science News with the headline, “The Original CRISPR: Before becoming a famous tool, the gene editor was a weapon in an unending microscopic war.”