STRANGE RELATIONS People, pandas, and even mushrooms perch as closer cousins to single-celled choanoflagellates than to other multicellular organisms.
The tree of life might seem like a stable design, appropriate for indelible ink. Plenty of people think so. An Internet search for “phylogenetic tattoos” turns up some showy skin art.
But the branches are shifting. Since a radial diagram based on 1990s genetics inspired a rush for tree-of-life tattoos, technical diagrams of life’s ancestral connections have been redrawn. And the simplified version of the tree of life memorized by schoolchildren for decades lags far behind what researchers depict today.
When Patrick Keeling at the University of British Columbia in Vancouver unveils a working scientist’s current diagram for his students, most have never heard the names of the major branches. “Kind of fun,” Keeling says.
In the new vision — based on increasingly sophisticated genetic analyses — people and other animals are closer cousins to single-celled choanoflagellates than to other multicellular organisms. Giant kelp that grow as wavering undersea forests off the California coast are closer relatives to single-celled plankton called diatoms than to multicelled red seaweeds or plants.

The old tree isn’t exactly wrong. The kingdoms that used to crown its top — plants, fungi and animals — still exist. But they’ve moved. In the new diagram, the tree’s former crowning glories shrink to mere side branches, three among hundreds, crowded by the vast diversity of complex single cells.
Biologists analyzing this treetop rarely use the word kingdom anymore. They talk of five or maybe seven bigger branches called supergroups. And the story of demoting kingdoms and introducing supergroups is far from over. A 2014 review noted five proposals for designating the most ancient stretch of supergroup branches, the bit that goes lowest on the new tree. A paper appearing in the Proceedings of the National Academy of Sciences in February described a new method for resolving this debate, and discussions continue.
Even as a work in progress, the new groupings offer benefits, such as help in searching for new medicines and insight into evolutionary puzzles such as the supposed wastefulness of sex. As the tree continues to morph, it will no doubt inspire new tattoos.
Story continues below graphic
Old school
Over time, drawings of the tree of life have changed to keep up with evolving ideas about how to categorize life.
In 1735, Swedish physician and botanist Carl Linnaeus published the first edition of his classification system for nature, incorporating three kingdoms: plants, animals and stones. (Stones never caught on.) In that early scaffolding, evolutionary relationships between organisms didn’t matter. Linnaeus was just striving “to reveal God’s creation to mankind in an orderly manner,” says archivist Gina Douglas of the Linnean Society of London. Charles Darwin wouldn’t nervously publish On the Origin of Species for another 124 years.
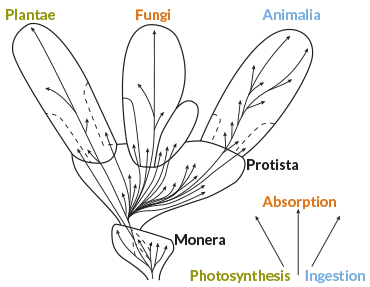
By 1969, ecologist Robert Whittaker openly waved aside some evolutionary distinctions when describing his ideas for kingdoms, in Science (see illustration below). His approach to the top of the tree (home to eukaryotes, which wrap their DNA in membrane-bound cell nuclei) would rule biology classrooms for decades. Yet his system is based only partly on evolutionary relationships.
Whittaker crowned his tree of life with three kingdoms of primarily multicellular eukaryotes sorted in large part by nutritional style: plants (capturing light energy), fungi (absorbing nutrients by contact) and animals (ingesting their food). He recognized a fourth kingdom of eukaryotes: the Protista. It was a hodgepodge of single-celled organisms, differing in forms and lifestyles but lumped together largely for convenience. He said as much, acknowledging that none of the classification systems he discussed “can be wholly satisfactory.”
Today many scientists hope for biological categories that consist of a single ancestral lineage and all its evolutionary descendants. A tree of life built this way “actually changes how you interpret a lot of stuff,” Keeling says. Much of biology rests on information gleaned from one organism and hypothesized to be true for its close kin. Identifying relatives is important. “If you’ve got the tree wrong, then you could be wrong about a lot of things,” he says.
The challenge of getting that tree right looms sequoia-high, since so much of the diversity of life lies in complex microbes still unknown to science. Out of about 150,000 kinds of marine plankton detected during the Tara oceans expeditions, as reported May 22 in Science, researchers could genetically identify about a third only as some eukaryote. They couldn’t place them in any known group.
Great idea, but
The early attempts to see what genetics could say about the tree of life seemed to work beautifully. But “science doesn’t go in a linear path,” says Andrew Roger of Dalhousie University in Halifax, Canada, who entered the field during the tumultuous 1990s.
Those first genetics-based trees of life for eukaryotes were built by comparing variations in the gene for small subunit ribosomal RNA across species. The result looked believable, with plants, animals and fungi on big branches at the top. Lower down, in the zone for more ancient branches, sprouted some oddball parasites such as Giardia (the bane of hikers who drink untreated water), sexually transmitted Trichomonas and tiny microsporidia, which attack many animals.
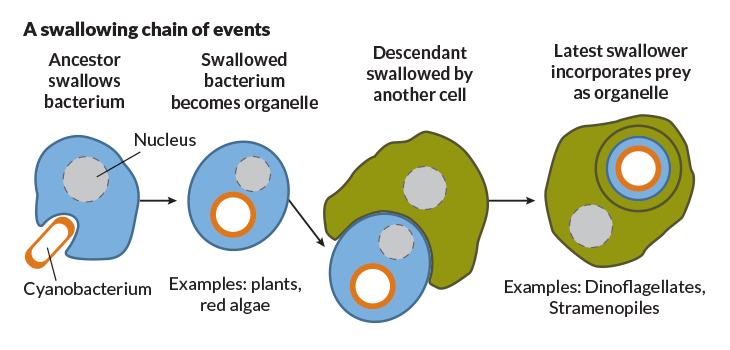
Researchers had begun to wonder if these bizarre parasites and their relatives could be living relicts of an early, pivotal time in eukaryote history. The organisms had no obvious mitochondria, the organelles that serve as the cell’s power stations. Perhaps the parasites had never had any mitochondria, Thomas Cavalier-Smith of the University of Oxford had suggested in 1983. This notion played off the idea championed by maverick biologist Lynn Margulis at Boston University in 1970 in The Origin of Eukaryotic Cells. She suggested that mitochondria came from a free-living microbe that some ancestor of eukaryotes had swallowed and put to work. Perhaps the parasites were relicts from before the Big Swallow.
To use these select parasites as glimpses of life before mitochondria “would be really, really interesting,” says Dalhousie biologist Alastair Simpson. “It was a lovely hypothesis. But then it all went to hell.”
One problem came when researchers expanded their analyses to look at more than one gene. Unexpectedly, says Roger, “we had different genes saying different things.”
The potential unraveling of the parasites-as-relicts hypothesis had a special zing for Roger. He was in the middle of a Ph.D. project based on the assumption that the parasites came from premitochondrial times. “It was one of those cases when you’re trying to work on the carpet that’s being pulled out from under you,” he says. “I thought it was pretty exciting.”
Yes, he said “exciting.” That view of eukaryote evolution based on analysis of a single gene “was in textbooks, and it was what many people had made their careers on,” he says. “It seemed like if it was going to fail, it was going to be something big.” He and his lab mates were going to be in on it.
Roger and his colleagues struck a blow to the old hypothesis by looking for genetic traces of bygone mitochondria lingering in the cell nucleus of select parasites. Even though mitochondria need some 700 to 1,000 genes to operate, only about 10 to 100 of them are inside the organelle itself. The rest reside in the cell nucleus and ship proteins to the mitochondria. The researchers detected some left-behind nuclear genes in supposed ancients, such as Giardia, Trichomonas and some microsporidia.
Various research groups eventually found the tiny, overlooked remnants of mitochondria themselves inside the fake relicts. The parasites looked very ancient only because their mitochondria had shriveled into hard-to-recognize bits.
Microsporidia, it turned out, were nowhere near being relicts. Examining a handful of genes, Keeling and other researchers demonstrated that the single-gene tree had put microsporidia in the wrong kingdom.
Microsporidia are “really nasty parasites, but fascinating,” Keeling says. Their tough spores enclose a coiled tube. To infect a victim, they pump inward such a rush of water that the pressure blows the top off the parasite and the tube shoots out like a harpoon. As much as 100 times longer than the spore, the tube-harpoon punches into its prey. The parasite cell nucleus and other internal parts are injected through the tube into the victim. Although they have this elaborate equipment for infection, microsporidia are simple cells with genomes that are “tiny, tiny, tiny,” Keeling says. Another misleading hint that they were ancient, simple creatures.
Keeling and his colleagues in the 1990s found evidence that the microharpooners didn’t belong at the bottom of the tree. They were fungi, a more recent and much more metabolically versatile branch of organisms. Before this revelation of misclassification, researchers accepted the simplicity of microsporidial cells by assuming that complicated traits hadn’t evolved in supposedly ancient, simple cells. After recognizing microsporidia as members of the fungal kingdom with all its elaborate metabolic tricks and lifestyles, biologists could see that microsporidia had been more complex at one time, but eventually lost fancy traits. “You have the exact opposite view of how they got to be the way they are, based on where they go on the tree,” Keeling says. “The tree matters.”
As evidence built that the supposed ancient parasites and their relatives weren’t so ancient, “smart people worked out that the molecular trees were just wrong,” Simpson says. The problem, now widely recognized, was the peril of “long-branch attraction.”
Lineages that change a lot end up as long-branch lines in an evolutionary tree. Younger branches that change fast can by chance develop similarities to genuinely old branches. In simple analyses, they end up closer than they should be at the base of the tree, as if they’ve “attracted” each other.
Supergroups
All the challenges to the first genetic tree of life “left us with a mess,” Keeling says. It became clear that the history of all eukaryotes cannot be reconstructed in any sensible tree based on one gene. “We ripped it all apart and put it back together. And it came together very differently.”
Instead of a crown of multicellular groups rising over relicts on long branches, the organisms “coalesced in a small number of very large groups,” he says.
The new arrangement, summarized in 2012 by Sina Adl of the University of Saskatchewan and colleagues in the Journal of Eukaryotic Microbiology, makes a fabulous mix of convergence and divergence. Animals are close relatives of fungi. Both are opisthokonts, along with some one-celled cousins. A Phytophthora potato pathogen, once famous as the “fungus” that caused famine in Ireland in the 1840s, is not a fungus at all. It belongs in the same supergroup as the giant kelp. Red and green seaweeds join plants in a distinct group called the archaeplastids.
These are big, deep-history groups, but don’t call them kingdoms. “Alastair is allergic to kingdoms,” Roger says of Simpson. The two collaborated on a 2002 paper enumerating supergroups instead.
Simpson wanted a term no one would treat as a formal rank. “ ‘Kingdom’ has such gravitas,” he says, not admiringly. Biologists assign kingdom rank based largely on subjective impressions, so he sees little information conveyed just by knowing something is a kingdom instead of some other rank in a hierarchy. To avoid the whole nonsense, he uses supergroup in a lighthearted nod to bands such as the Traveling Wilburys, which united (briefly) Bob Dylan, George Harrison and three other independently famous musicians.
Sequential swallowings are a big part of eukaryote history. Engulfers were themselves engulfed and in turn engulfed yet again. “Like matryoshka dolls,” Keeling says.
Some genetic evidence now links three big groups to a single ancestral engulfment of chlorophyll c, Simpson says. Stramenopiles, with giant kelp and potato pathogens, now join alveolates and rhizaria (even though they have no chlorophyll c) as what’s often abbreviated as SAR. Other chlorophyll c carriers are still under study, so the bold hypothesis might someday turn out to be right. “The really cool thing is, we still don’t know the answer,” Simpson says.
This and other eukaryotic mysteries may resolve more easily as geneticists refine a technique for deciphering DNA from one individual cell. Nature Methods called it “method of the year” for 2013.
The technique’s possibilities intrigue researchers because single-celled species can be difficult or impossible to grow in the lab. Roger lovingly describes radiolarians, some of which build skeletons of strontium sulfate while others manage to eat multicelled animals. “Just beautiful organisms,” he says, but “they’re a real pain actually to work on.” Single-cell DNA sequencing has helped in the study of bacteria, and papers here and there report results for eukaryotes. The technique revealed an interplay of chemistry between a termite-gut dweller and its own live-in spirochete bacteria in work published May 15 in the Proceedings of the National Academy of Sciences.
Practicalities and beyond
Reconstructing all this evolutionary history can have practical implications. Early in Keeling’s career, he investigated the peculiar discovery that malaria parasites have remnants of organelles that once carried chlorophyll. “Why would malaria have a chloroplast? They live in the dark,” Keeling says. The researcher, Iain Wilson, who originally proposed the notion was dismissed for a long time as ridiculously wrong. But evidence grew, and today scientists agree that a certain small bit in the parasite left over from a photosynthetic past still has some function in the dark. That finding helps explain the effectiveness of some formerly mysterious treatments for malaria: The drugs attack chloroplasts.
A realistic tree of life can also change the way researchers look at more theoretical questions, such as how in the world such a supposedly wasteful and irrational process as sex evolved. Only one of two partners can bear young, halving productivity from the start. Or so goes a long-running thread of research and debate (SN: 2/14/09, p. 16).
Yet that furor is so multicellular. Simpson and Roger along with philosopher Maureen O’Malley of the University of Sydney in Biology & Philosophy in 2013 pointed out that the basics of sex must have evolved long before birds, bees or anything else multicellular.
In single-celled organisms, reproduction does not require sex, nor does sex always produce offspring. Two ciliate cells can meet, mingle genes and remain only two cells. The eukaryote tree abounds with single-celled organisms practicing the basics that combine to make multicellular reproduction possible. The alleged wastefulness of sex is really a question of why the peculiar form of sex tied to multicellular reproduction persists in a few outliers.
Useful as a good tree of life can be, Keeling objects to the idea that eukaryotes should be appreciated just for their utility. People enjoy animals and plants for their own sake. “A giraffe won’t save your life, but people like giraffes,” he says. To him, one-celled eukaryotes are Serengeti charismatic, just smaller. Some species have a structure — similar to camera-like animal eyes — with a cornea-like outer covering derived from mitochondria and a light-sensitive inner cup from an engulfed red-algal organelle, Keeling and colleagues reported July 9 in Nature. Several large dinoflagellate species hunt despite living inside a rigid shell. They push a stomachlike structure out a hole in the shell and digest prey larger than they are. Other single cells grow delicate, multipointed outer casings that eventually wash ashore, covering beaches with millions of miniature stars.
Knowing that the living world has so much invisible variety can change a person’s perspective, says Fabien Burki, who works with Keeling at the University of British Columbia. The supergroup tree offers the little back-of-the-neck shiver-thrill of realizing that every tomato patch, termite gut or beach bucket of seawater holds life much vaster and stranger than imagined. Says Burki: “It’s like the astronomers discovering there are planets around other stars.”
This story appears in the August 8, 2015, issue with the headline “Strange Relations: Schoolroom kingdoms are taking a backseat to life’s supergroups.”