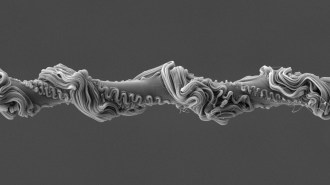
Microbiologists took 12 years to grow a microbe tied to complex life’s origins
Scientists are hopeful that an archaean may help answer how multicellular life evolved
Scientists spent over a decade growing this microbe, which they named Prometheoarchaeum syntrophicum, from deep-sea sediment.
M.K. Nobu and H. Imachi
Cramped in a small submarine 2,500 meters below the Pacific’s surface in 2006, microbiologist Hiroyuki Imachi scanned the ocean floor for signs of microbial life.
As the sub drifted over the bottom of Japan’s Nankai Trough — a hotbed of understudied microbes living off methane bubbling up from tectonic faults — Imachi spotted a nest of small clams against a whitish microbial mat, suggestive of an active methane seep below. The submersible’s robotic arm plunged a 25-centimeter tube into the blackish-gray sediment to retrieve a core of muck.
It would take another 12 years of lab work for Imachi and colleagues to isolate a prize they hadn’t even set out to find — a single-celled microbe from an ancient lineage of Archaea, a domain of life superficially similar to bacteria. That find could help biologists reconstruct one of life’s greatest leaps toward complexity, from simple bacteria-like organisms to more complicated eukaryotes, the enormous group of chromosome-carrying creatures that includes humans, platypuses, fungi and many others.
“Patience is very important in doing successful science,” says Imachi, of the Japan Agency for Marine-Earth Science and Technology in Yokosuka. He and his colleagues published their findings in the Jan. 23 Nature, to enthusiastic acclaim from fellow microbiologists. “I’m very lucky.”

Many scientists think an unusual meal kicked off the evolution of more complicated cells about 2 billion years ago. An ancient archaean, the theory goes, gobbled up a bacterium that, instead of being dinner, sparked a symbiotic relationship in a process called endosymbiosis (SN: 6/8/74). Eventually, the bacterium evolved into mitochondria, the energy-producing cellular structures that fueled the rise of complex life.
Living remnants of ancient archaeal lineages persist in some of Earth’s most extreme environments, and scientists are exploring these microbial hot spots for clues about the ancestor of all eukaryotes. One such environment is the deep-sea floor. Despite making up about 65 percent of Earth’s surface, biologists have only a faint picture of the microbial multitudes that thrive there. Genetic sequencing of dredged up mud has given biologists one way of studying these communities of bacteria and archaea uniquely adapted to the cold, oxygen-less deep. But genes can reveal only so much.
So scientists seek to grow cultures of microbes in the lab to study what these organisms look like and how they behave. But extreme microbes present unique challenges. Simply plating these organisms on a petri dish, providing nutrients and waiting for growth hadn’t ever worked — possibly because scientists weren’t effectively re-creating the microbes’ extreme environment, says Masaru Nobu, a microbiologist at the National Institute of Advanced Industrial Science and Technology in Tsukuba, Japan, who joined Imachi’s project after it started.
So Imachi, Nobu and their colleagues tried to re-create a methane seep in the lab, drawing inspiration from a bioreactor used to treat municipal sewage. The team pumped methane gas into a meter-tall cylindrical chamber, kept at 10° Celsius and stacked with polyurethane sponges that mimic porous deep-sea sediment. A slow, steady flow of artificial seawater kept the sponges saturated.
The team then watered down a clump of mud from the Nankai Trough sediment core, sopped up the slurry with the sponges, stacked them in the reactor — and waited.
“There was a lot of nervousness,” Nobu says of that time in December 2006. “We didn’t know if we’d get what we wanted.”
Each year, the researchers sequenced genes of microbes in the sponges. After a volatile first couple of years, the microbial community began to stabilize and grow. “Most of the organisms that were active in the reactor were organisms that were actually active in the natural environment,” Nobu says. With a stable community of thousands, if not tens of thousands, of different kinds of microbes to draw on, the team could try to pick out and grow individual strains.
Samples from the reactor were placed into 200 glass bottles, each filled with a different energy source and cocktail of antibiotics to weed out bacteria and allow different archaea to grow.
The team had its first eureka moment in 2012, detecting an archaean new to science that they called MK-D1 in very low numbers amid numerous bacterial strains in one bottle. But each time the team tried to isolate the archaean in a new bottle, the microbe just wouldn’t grow. Months of trial and error followed. “It was really frustrating,” Nobu says.
Then, the researchers had an idea: Perhaps the microbe was actually growing, but at a slow pace, a result of its deep-sea home. “It’s very cold down there, there’s not a lot of energy,” Nobu says.
So the team measured growth with a more sensitive technique, called quantitative PCR, that can quantify abundance from whiffs of DNA. Sure enough, MK-D1 was there and growing, just more slowly than any other single-celled microbe ever cultured. E. coli, for instance, can replicate itself in about 20 minutes. MK-D1 takes two to three weeks to divide.
“No microbe we knew about grew this slowly,” Nobu says. “Understanding this was a revelation.”
Meanwhile, another archaea discovery in 2015 rocked the world of microbial ecology. A new group dubbed Asgard archaea had been discovered from genetic material dredged up from a hydrothermal vent in the Arctic Ocean. Asgards have many eukaryotic genes, leading some scientists to argue that Asgards are the closest living relatives of ancient archaea that may have given rise to all complex life on Earth (SN: 12/15/15).
Sign up for our newsletter
We summarize the week's scientific breakthroughs every Thursday.
Imachi and Nobu were stunned when DNA evidence confirmed that they’d unwittingly spent the last nine years cultivating their own Asgard, MK-D1. If it could be isolated, Imachi’s team would be the first to actually glimpse a living member of this exciting but mysterious group.
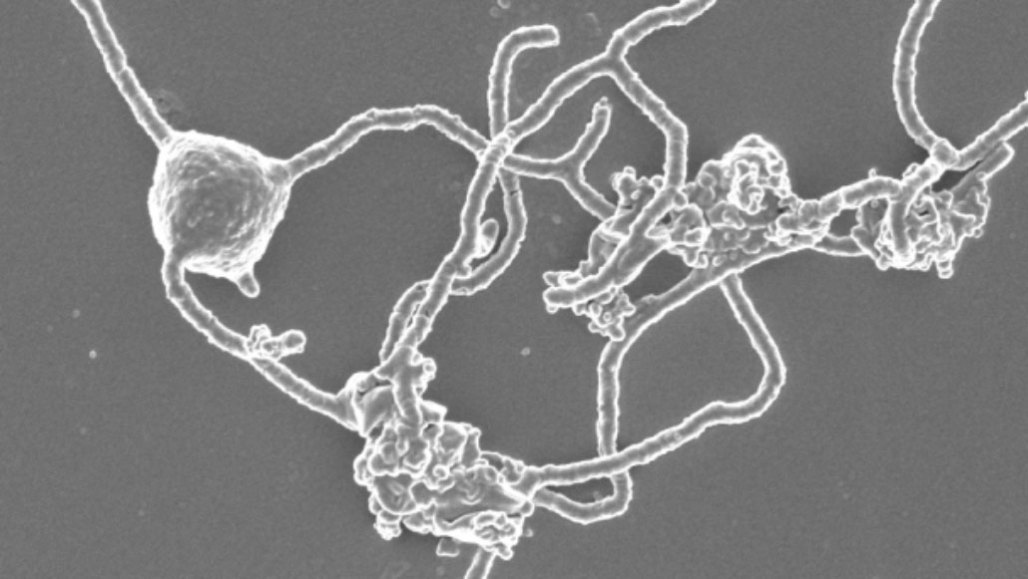
The researchers finally got a stable culture of MK-D1 to thrive — with a bacterial partner that it needs to survive — and in 2018, took their first look under a microscope. The neat, tiny spheres seen at first seemed unlikely to be the sort of thing that may have begotten complexity. But over months, the microbes grew odd, tentacle-like protrusions. Imachi “initially thought the sample had been contaminated,” he says. But the observation was sound, prompting the researchers to propose a model for how these tentacles might have ensnared other microbes — a probable first step in endosymbiosis.
The team gave MK-D1 a proper name, Prometheoarchaeum syntrophicum, after the Greek god Prometheus who, the myths say, introduced fire to humanity. Much remains to be learned about P. syntrophicum and what, if anything, it can tell us about our origins. In the meantime, Imachi is still sifting through the microbes in his reactor.
As he puts it, “uncultured microbes are waiting.”