Unnatural selection
Chemists build proteins with parts not in the typical toolkit
Amino acids are the Legos of life — tiny bricks that snap together, forming the proteins on which every function of life depends. With rare exceptions, cells choose from just 20 kinds of Legos. But this is enough for human cells to assemble the more than 1 million proteins they need to function.

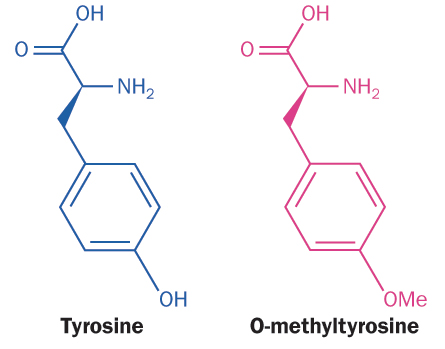
A couple of decades ago, a few scientists decided that they wanted to play with more Legos. It began as an exercise in academic curiosity, a way to ask some of the Big Questions about life: Why just 20 amino acids? Why those 20? The researchers began to build artificial amino acids in the laboratory — just to see what cells would do with new construction material, and where the exploration would lead.
Today, scientists have created more than 70 of these “unnatural amino acids” and are using them to reboot the protein-making machinery of bacteria, yeast and even mammal cells — all of which seem to welcome extra choice in their protein assembly. Given the success so far, at least two U.S. biotech companies are now using unnatural amino acids to mass-produce proteins previously unknown in nature, aiming to make new drugs that may one day treat cancer, multiple sclerosis or other diseases.
And drugs could be just the beginning. Artificial amino acids could take paths that no one can yet foresee, just as the inventors of nylon in the 1930s could not envision the day when their new material would lead to credit cards and water bottles, says Andrew Ellington, a biochemist at the University of Texas at Austin. “This is going to be a very powerful technology,” he says. “We can recast the very chemistry of life.”
The protein challenge
Chemists have been making new molecules for centuries, but mostly creating compounds that are small and easy to manipulate. Synthesizing a protein is a formidable challenge — so much so that the 1984 Nobel Prize in chemistry went to the man who mastered it.
Relatively speaking, proteins are enormous: Compared with a typical drug, a single protein could easily be 500 times larger and have mind-blowing complexity. Once constructed, a protein also has to contort itself into a unique three-dimensional shape; without this precise chemical origami, a protein is biologically useless. (The disease cystic fibrosis, for instance, occurs when just one of the body’s proteins gets misshapen.)
And once created, proteins are not all that stable: A protein that gets too hot or too cold will radically change its state and function — a property that allows humans the benefits of cooking and distillation. In short, says Peter Schultz, a chemist at the Scripps Research Institute in La Jolla, Calif., “making a protein takes a lot of chemical steps.”
In the 1980s, Schultz and others started brewing proteins and inserting artificially created amino acids by hand. These first proteins were painstakingly slow to make, so in the late 1990s researchers started enlisting bacteria. A bacterial cell could incorporate unnatural amino acids into its own proteins. But doing so meant overriding the cell’s genetic programming. “You’re actually changing the fundamental way genes are read,” says Chang Liu, a chemist and fellow at the Miller Institute for Basic Research in Science at the University of California, Berkeley.
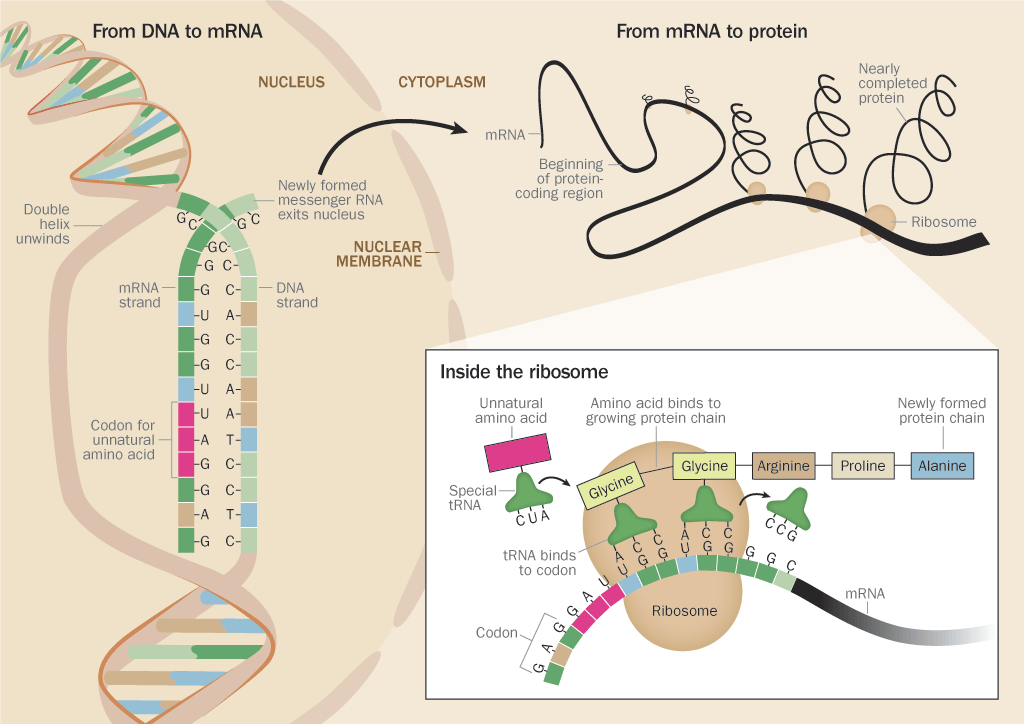
The genome, or repertoire of genetic material encoded in the DNA of each cell, contains the instruction booklet for each protein. A gene guides a cell’s step-by-step protein creation, dictating which amino acid to use and where to use it. The language of DNA is spoken in chemical bases — adenine, guanine, cytosine and thymine. Each combination of three bases codes for a certain amino acid. When need arises for a protein that has to perform a certain task — say, one vital for muscle movement or breathing — the code is transcribed by RNA, DNA’s sister molecule.
RNA transfers the code as three- letter “words” written with adenine (A), guanine (G), cytosine (C) and uracil (U), the base substituted for thymine in RNA. These four bases have 64 possible three-letter combinations. For example, UGG is the code for the amino acid tryptophan. UCC is the code for serine. So an RNA sequence of UGGUCC would mean “connect tryptophan, then serine.”
Writing new genetic codes for new amino acids posed one fundamental problem: All 64 three-letter words were spoken for. So Schultz and his colleagues zeroed in on one of the few combinations that did not correspond to an amino acid — the sequence UAG. UAG is a genetic punctuation mark; as the protein factory line works its way down the RNA, reading the code, UAG means “stop, the protein is finished.”
The Scripps scientists took advantage of UAG’s lack of specificity, and reprogrammed the combination to correspond to one of their new amino acids. “We’re not changing the DNA,” Schultz says. Rather, the researchers give existing DNA new meanings.
The researchers first announced success in 1999 using the bacterium E. coli (SN: 6/3/00, p. 360), and in the decade since, Schultz and others have used UAG reprogramming to get artificial proteins not only out of bacteria, but also yeast and the cells of mammals. This February, in the journal PLoS One, Schultz’s team described getting the tuberculosis bacterium to accept an unnatural amino acid, one of the first important human pathogens to do so. Being able to manipulate the proteins of the tuberculosis germ might allow researchers to better study the organism and develop new vaccines.
Rewiring UAG has its limits, however. For one thing, scientists can still add only one new amino acid at a time, maybe two. (In addition to UAG, two other codes signal “stop.”)
How to trick nature
Caltech chemist David Tirrell has devised a different method to get artificial amino acids into proteins. He doesn’t reprogram a cell’s existing code, but substitutes an unnatural amino acid that closely resembles a natural one. “The cell thinks it’s putting in what it’s coding for,” he says. It would be like swapping a red Lego for a green one with the same size and shape. If the red Lego isn’t around, the cell will grudgingly pick up the next closest thing. This method allows a much wider use of unnatural amino acids, but also has certain drawbacks — as you might not always want a green Lego in every spot that calls for a red one.
Molecular geneticist George Church of Harvard University and colleagues described last year in Nature still another way to get around genetic programming. His method has the potential to rewrite many three-letter codes at once. Just as different words can signify the same thing — cash, bucks and moola all mean money — most amino acids use more than one code. For example, the sequences GUU, GUC, GUA and GUG all code for the amino acid valine. Church’s laboratory has developed technology that can erase duplicate codes — say, GUU — and rewrite them for a custom amino acid. “We can’t invent a new set of three letters,” Church says, “but we can knock out the ones we want.” Once the code loses its original meaning, the scientists are free to rewrite it as they choose.
Now that researchers have shown that altering the genetic code is possible, they must ask themselves whether doing so serves any purpose. After all, the current code is the product of nearly 4 billion years of trial and error. Wouldn’t the best amino acids and the best proteins be left standing?
Not necessarily, Church says. For one thing, the current amino acids are the result of natural selection, but humans no longer live in a natural world. The use of unnatural amino acids might allow proteins that can function in totally man-made environments, such as oil refineries. After all, some of the odd forms of life that use a naturally occurring 21st amino acid, one-celled microorganisms belonging to the group Archaea, can exist at the extreme pressures and temperatures of hydrothermal vents on the deep ocean floor.
Many researchers are also hoping that unnatural amino acids can lead to new ways of treating disease. An artificial amino acid can provide an extra “chemical handle” on an otherwise normal molecule, says Ho Sung Cho, chief technology officer of the California biotech company Ambrx (founded in part by Schultz, who is a stockholder). If the molecule has a spot for something to dock onto, researchers can snap on whatever chemical accessory they wish. “It’s like adding a USB port,” Cho says.
The company is already testing proteins with extra amino acids in human clinical trials. For example, people who must take growth hormone now require daily injections of the medicine because the protein degrades quickly in the body. Ambrx has added a 21st amino acid to its clinical preparation of growth hormone, and says the modified hormone remains biologically active for days. The compound is still undergoing testing in people. But at a 2008 meeting of the International Congress of Endocrinology, Ambrx scientists reported on a study of 22 volunteers that found that an injection of the new version can remain biologically active for at least a week.
The company is working on other molecules incorporating unnatural amino acids, including some that could potentially be used to treat multiple sclerosis and diabetes. Another possible chemical attachment is a toxin hooked to an antibody, which zeroes in on a cancer cell to attack it. This method has long been attractive to cancer researchers, but natural proteins have often been too unstable to reach their target before releasing the toxin, Cho says. The hope is that an unnatural amino acid would allow the molecule to hold together longer.
Drug development
Meanwhile, the Seattle biotech firm Allozyne (founded in part by Tirrell, and using his approach), employs the amino acid substitution method to build a protein that might treat multiple sclerosis. As with the growth hormone modifications, the addition of an unnatural amino acid may help the molecule — in this case, the MS drug interferon beta — last longer in the body, says Ken Grabstein, Allozyne’s chief scientific officer. The drug is in safety tests with volunteers who have multiple sclerosis, though the results have not been released.
Like all biologic drugs, those with artificial amino acids raise safety questions: Will the new protein provoke the body’s immune system to attack? So far, Grabstein says, animal studies indicate that the body will accept the new drugs, but this and other questions of safety will not be answered until larger studies are underway.
Potential uses for artificial amino acids go beyond new medicines, such as getting unusual glimpses of life’s hardware. At Caltech, Tirrell is flagging proteins with artificial amino acids as the molecules are made, providing the ability to see what product a cell is making at any given time. It’s like having a live video feed of protein synthesis. “This will help us understand how a cell is responding to its environment,” he says.
Schultz hasn’t abandoned the original mission that intrigued him — learning what, if anything, makes the 20 amino acids that nature provides so special. It’s possible that those 20 are the optimal combination. It’s also possible that those are, as the late biologist Francis Crick once described it, part of a “frozen accident” that left people with their current genetic code. Perhaps nature settled on those 20 simply because they offered the first combinations that sustained life at all, not necessarily the combinations that work best. Should other forms of life on distant planets use amino acids, the selection might look different.
Scientists know that they are experimenting with some of life’s most basic elements. “Is there a reason we shouldn’t have unnatural amino acids?” asks Church. “Theoretically, you could say that we could make an organism that would outcompete something in the wild.” In other contexts, foreign life has entered natural environments with catastrophic consequences: Asian carp in North American waterways, European rabbits in Australia or kudzu in the American South. Exotic organisms have a history of causing native surroundings to collapse.
Church doesn’t think that could happen easily or accidentally — for one thing, left to its own devices, an organism designed to function with an extra amino acid wouldn’t survive if that extra component weren’t around. Nonetheless, Church says, “we should do experiments to see what the consequences would be.” He plans to undertake those in the next few months, by growing an E. coli that requires unnatural amino acids and mixing it with natural strains in laboratory dishes. Most likely, the strain with unnatural proteins won’t survive, even if its extra amino acid is available, but Church won’t know for certain until the experiment is done.
Schultz already plans to see whether life with 21 amino acids fares differently than it does with the usual 20. By next year, he hopes to be able to breed a mouse whose cells can readily use 21 amino acids in daily life. Experiments on a smaller scale have suggested that organisms with 21 amino acids can, in a kind of laboratory-simulated natural selection, develop proteins with extra functions that give them an evolutionary advantage over proteins with 20.
“The 20 amino acids are ideal for proteins that support all life,” says Thomas Magliery, a biochemist at Ohio State University in Columbus. “That doesn’t mean they are ideal for all possible things.” Scientists have spent the past decade making sets of Legos never previously imagined. It’s time now to see what these new blocks will build.