Here’s the poop on getting your gut microbiome analyzed
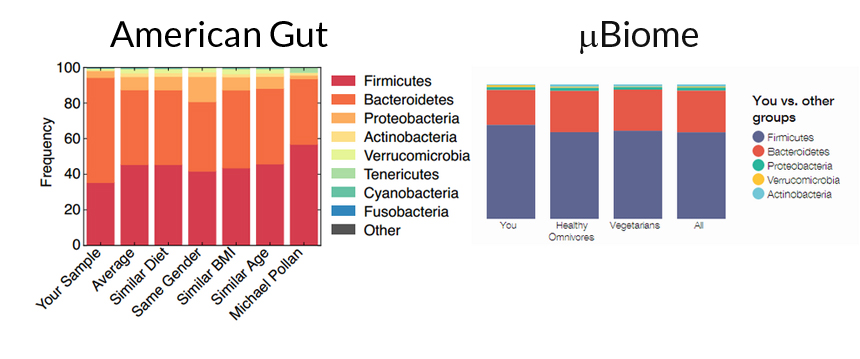
I asked two different companies to analyze my gut microbiome. American Gut (left) gave nearly opposite results to those from uBiome (right) with respect to the major phyla of bacteria in a duplicate sample.
American Gut, uBiome
Guest post by Tina Hesman Saey
I donated my used toilet paper to science. The act wasn’t a prank or a weird protest; it was an effort to discover what microbes are living in my intestines.
Those microbes in and on your body include bacteria, which outnumber your own cells 10 to 1. Together with with fungi, archea, viruses and other single-celled organisms, they are known collectively as the microbiome, and affect our health in myriad ways, both good and bad. As a result, microbiome research has become one of the hottest topics in life science research in the past decade or so. Little did I know when embarking on a personal quest to learn more about the microbes I house that the field is also about as uncharted and free-wheeling as the Wild West.
Prior to 2013, about the only way you could get a genetic analysis of your own microbiome was to join a study. Even then, your data would be anonymous and you probably would never learn what the researchers found in your gut, mouth, vagina or nose or on your skin. But last year two services, American Gut and µBiome (pronounced you-biome), launched through the crowdfunding website Indiegogo. Those companies will sequence your microbiome for just under $100 and send you a report of the results.
I signed up for both. For µBiome, I swabbed my mouth to learn more about its inhabitants, and I rubbed giant Q-tips on my forehead so American Gut could tell me what lives on my skin. And I sent fecal samples to both places. I gave the services as close to duplicate samples as I could, swiping big cotton swabs over the same section of used toilet paper — per their instructions — within seconds of each other. I wrote about the preliminary results µBiome sent me, but it took several more months to get results back from American Gut. I’m still waiting for the skin results.
When the second set of gut results came in, I was shocked. The results didn’t match at all with µBiome. In fact, they were almost complete opposites of each other in regard to the proportion of Firmicutes and Bacteroidetes that make up my fecal microbiome. Firmicutes and Bacteroidetes are the two major phyla of bacteria found in the human gut, and their proportions can say a lot about what your diet and health may be.
Here is a look at the top 10 bacterial genera identified by the two services:
American Gut |
µBiome |
||
| Genus | Percent of sample | Genus | Percent of sample |
| Bacteroides | 46.92 | Bacteroides | 20.3 |
| Ruminococcus | 4.78 | Faecalibacterium | 12.2 |
| Parabacteroides | 2.51 | Blautia | 9.28 |
| Faecalibacterium | 2.37 | Ruminococcus | 4.81 |
| Blautia | 2.21 | Streptococcus | 3.67 |
| Sutterella | 1.72 | Roseburia | 2.05 |
| Lachnospira | 1.29 | Dorea | 1.48 |
| Akkermansia | 1.23 | Akkermansia | 1.42 |
| Ruminococcus* | 0.94 | Alistipes | 1.39 |
| Streptococcus | 0.92 | Parabacteroides | 1.21 |
* Denotes Ruminococus from the family Lachnospiraceae. The more abundant Ruminococus genus is in the family Ruminococcaceae.
I thought I’d see if the Twittersphere was as puzzled as I was by these results:
How does my microbiome compare to yours. Results from @americangut and @uBiome pic.twitter.com/5VMpkNW72V
— Tina Hesman Saey (@SN_Saey) April 3, 2014
A blogger who follows my Twitter feed was disturbed by the discrepancies and called for American Gut and µBiome to explain how the same sample could produce such wildly different results.
It turns out that I’m not the only one to notice problems with the companies’ fecal microbiome analysis. One blogger found differences between the microbes taken from two different parts of the same, uh, sample. It’s worth noting that American Gut and µBiome agreed on the microbe mix in one portion of her poo, but she only sent the other section of the sample to American Gut. That makes it very hard to judge if the same fecal sample can differ widely or if something in the sequencing and analysis process gave very different results.
So what could have gone wrong with my microbiome? Perhaps the samples weren’t collected right. According to American Gut’s sampling instructions, too much brown stuff can interfere with the methods the scientists use to break open bacteria and pull out the DNA inside. Too little and they might not get an answer at all. I thought I had used Goldilocks-like precision, and that one — or maybe both — of the sequencing services must be wrong.
Another possibility: corrections to the data may differ between companies. Another blogger, who is a bioinformatician, got different results than American Gut reported to him when he used his own software to analyze the raw data. It turns out that some bacteria grow while in the mail and can take over the sample, so American Gut corrects the reports it sends to participants to account for that overgrowth.
In the end, I thought I’d go right to the experts for the straight poop. I approached Jessica Richman, one of the cofounders of µBiome, and Rob Knight, one of the leaders in the microbiome field. His lab group runs the American Gut project as part of their quest to learn how diet, lifestyle, geography and other variables influence the microbiome.
They both gave me several technical explanations for what might have skewed my results. Richman offered a variety of possibilities in a blog post published on the company’s web site. Her number one candidate is the DNA extraction technique – the very one I was so careful not to interfere with by getting too much poo on the swab. It seems that the services use different kits to bust open bacteria and get at the genetic goodness inside. Other researchers have noticed that the way DNA extraction is done can make a big difference in the results. So µBiome took another look at my data and adjusted it to account for different extraction methods and came up with a result somewhere in the middle of the original two profiles — but still closer to µBiome’s original analysis.
But DNA extraction is not the only thing that could go wrong. It seems that every step of the process — from how you collect the sample through the computer programs used to analyze the DNA data — is a potential culprit. Or as Knight puts it, “All sorts of unlikely things are possible, and finding out which one is true is difficult.”
This has been a major headache for people in the microbiome field. Different labs each using their own protocols can arrive at widely varying answers to essentially the same questions, such as what mix of microbes is associated with irritable bowel disease or colon cancer.
For Rashmi Singha, an epidemiologist at the National Cancer Institute, the lack of reproducibility between studies was frustrating. “To me it seemed like cowboy country. It needed to have some kind of order.”
Together with Knight and 14 other well-established microbiome labs, Singha instituted the Microbiome Quality Control project to bring some consistency to the field. The labs are analyzing a variety of fecal samples to see if they can come up with the same answers about what’s in them. These include fresh or freeze-dried feces and some microbe mixes that Singha had Emma Allen-Vercoe at the University of Guelph in Canada cook up in an artificial colon called the Robogut. The Robogut can produce a liter of feces or more at a time (which is worth a blog post of its own). The point is, scientists are trying to find ways to standardize microbiome studies so that they can directly compare results. They don’t yet have the answers, but they will take the first steps toward figuring it out at a workshop this fall.
As for me, I’ve had my microbiome sequenced twice, but I still don’t really know what’s in there. The comparison data on the insanely fun µBiome results viewer assure me that the abundance of my various and sundry bacteria are mostly within normal parameters. According to my American Gut microbiome portrait, I fall pretty solidly near the center of people with a Western-style diet and the American Gut population itself (the big dots are me).
#americangut 4/4 pic.twitter.com/F5tLuSIhzl
— Tina Hesman Saey (@SN_Saey) April 3, 2014
Knight says that tells me something about my microbiome: “You can feel relatively secure that you’re in the distribution of healthy Western adults.”
When I asked for a redo on the microbiome sequencing, Knight said I, and science, would be better served if I get my samples into the next round of the Microbiome Quality Control project.
Maybe the next time I hand over my used toilet paper I’ll get a clearer picture of what’s really up with my microbiome.