Michael Phelps, one of the greatest swimmers of all time, propels himself forward by hurling water behind his body. If he were the size of a bacterium, though, that strategy wouldn’t make much of a splash. In a microworld Olympics, Phelps would go home medalless.
At tiny scales of 10 micrometers and below, life is largely conducted as if in a thick fluid, where every motion is immediately dampened by the highly viscous muck. Here, where water seems to take on the consistency of honey, the coasting inertia that helps carry Phelps through the water is simply nonexistent.
“It’s like looking at a completely different world,” says Piotr Garstecki, a physicist at the Institute of Physical Chemistry at the Polish Academy of Sciences in Warsaw.
Many microbes spend their whole lives swimming, and it’s not to win Olympic gold. They make their way through the thick morass to find food, locate mates and seek out or avoid light. But exactly how some of them manage such successful strokes has been a mystery, even though the critters swim right under (and in) researchers’ noses.
Recently, though, scientists have discovered some of the microswimmers’ tricks. Biophysicists and mathematicians have devised new equations to describe fast, efficient, small swimmers such as Spiroplasma bacteria, which move by propagating kinks in alternate directions along their spiral-shaped bodies. Other scientists are revealing more about the coordinated waltzes of algae and the gyrations of bacteria.
Understanding microorganisms’ swimming styles may shed light on infection control (stopping swimmers may halt infections), reproduction (sperm are some of the best swimmers around) and ocean ecology. And insights from this microworld have led other researchers to begin designing artificial microswimmers — tiny machines that may one day be used for tasks like rooting out plaque from clogged arteries or ferrying drugs to precise locations in the body.
“The most exciting idea is that understanding how microbes move can help us mimic that machinery and build small machines,” Garstecki says.
That aim has attracted a diverse crew of researchers. Microbiologists, of course, study swimming at tiny scales, but increasingly so do physicists, mathematicians, roboticists and microfluidics experts, to name a few. A special section in the May 20 Journal of Physics: Condensed Matter is devoted solely to the non-intuitive physics of microswimming.
“A lot of physicists realized that lots of questions have not been answered,” says Raymond Goldstein, a biological physicist at the University of Cambridge in England. This realization, added to new experimental tools and better modeling, has prompted an exciting resurgence in the field, he says.
Living without inertia
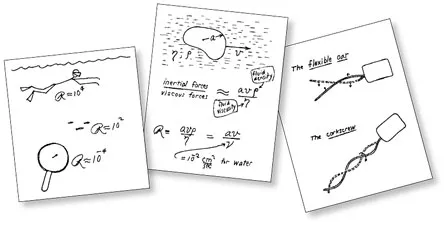
Scientists interested in microorganisms’ strokes have a very important number to contend with. Swimmers of all sizes must deal with a dimensionless value called the Reynolds number — the ratio of inertia (an object’s resistance to change in speed, whether faster or slower) to viscosity. Phelps has a high Reynolds number in the pool, exceeding 1 million, meaning that his strokes create sufficiently powerful inertia to carry him through the water, and viscosity is not overwhelmingly important. On the other hand, an E. coli bacterium swimming through the exact same water has a Reynolds number of about 0.00001. Inertia is virtually nonexistent, and viscosity is paramount. Under these conditions, when a bacterium stops moving an appendage, the fluid around it stops almost instantaneously.
“Motion at low Reynolds number is very majestic, slow and regular,” the late physicist E.M. Purcell said in a famous 1976 lecture on the physics of micro-organisms. “If you are at very low Reynolds number, what you are doing … is entirely determined by the forces that are exerted on you at that moment, and by nothing in the past.” In the talk, later published in the American Journal of Physics, Purcell made use of a large vessel of corn syrup to demonstrate the high viscosity that characterizes life on a tiny scale.
A special challenge of moving around in corn syrup is that reciprocal motions, like a scallop opening and closing its shell, yield no net movement. The movement achieved by the scallop opening, the first half of the stroke, would be exactly undone in the second half as the scallop slams shut. Similarly, a fish moves by swishing its tail back and forth, but if it lived at a low Reynolds number, the motion attained by a swish in one direction would cancel out the motion gained from the previous swish. To swim, Goldstein says, microorganisms “have to move an appendage in such a way that if you record it and play the movie backwards, you can tell which way is backwards.”
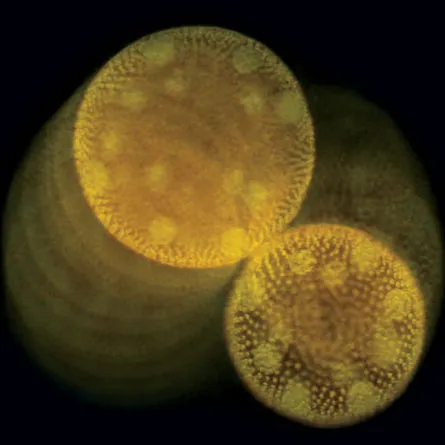
One of the best known examples of a tiny swimmer’s successful nonsymmetrical motion comes from E. coli’s stiff, corkscrew-shaped flagellum. As it rotates, this appendage pushes the bacterium’s body ahead and the liquid behind. Still other microorganisms, such as the Volvox algae, are covered in flexible, undulating flagella whose coordinated waves add up to net motion.
“Something that strikes me is the variety of different kinds of tiny little machines that these bacteria — and also archaea — have invented for movement,” says Mark McBride, a molecular geneticist at the University of Wisconsin–Milwaukee. In 2008, McBride coauthored a review paper in Nature Reviews Microbiology titled, “The surprisingly diverse ways that prokaryotes move,” which described “swimming, swarming, gliding, twitching or floating.”
Waltzing through honey
New work suggests scientists can add another movement to the list — dancing.
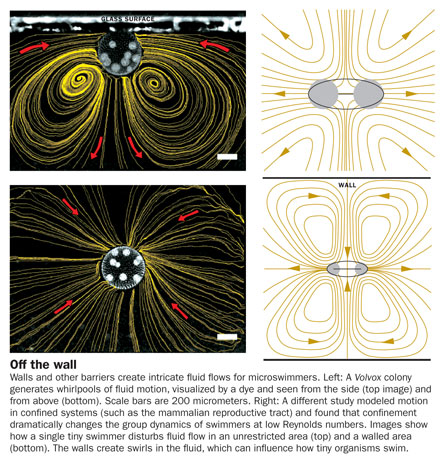
In addition to the honey consistency of life at low Reynolds numbers, all tiny swimmers must contend with powerful influences created by fellow swimmers and walls or other edges. “You can get interesting hydrodynamic effects and long-range interactions that are particularly strong,” Goldstein says.
These effects are at play in the unusually robust interactions of small (around 500 micrometers in diameter) spherical swimming colonies of Volvox algae, which operate at a Reynolds number around 0.03, Goldstein and his colleagues reported online April 20 in Physical Review Letters.
Viewed through a microscope, colonies of the algae perform a slow, clockwise waltz — in which the colonies circle each other — and a jumpier, quicker-paced minuet. These stable, paired dances are caused by the undulating flagellar motion of each algal colony and by the peculiar fluid flows near the glass coverslip, where the Volvox prefer to swim.
“We saw that they would linger around each other. They orbit around each other in miraculous dynamics — like the planets around the sun,” Goldstein says. He thinks that the dancing may enhance the chances of fertilization during the algae’s sexual phase. Sperm packets released by male Volvox may find their targets more quickly because of the dancers’ proximity. “The collective behavior can influence their life,” Goldstein says.
Kinky locomotion
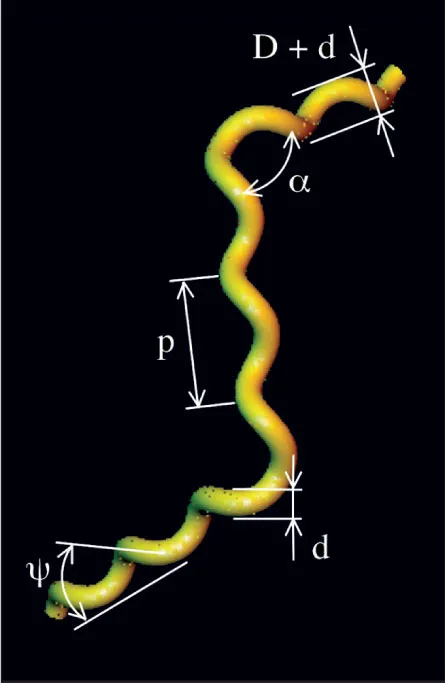
Edge effects and long-range splashbacks are everyday hurdles for microswimmers, who cut through the goo with ingenious tricks. One elegant design is seen in Spiroplasma. Tiny even by bacteria standards, Spiroplasma propels itself by propagating kinks through its corkscrew-shaped body. The kinks look like those formed in a phone cord twisted to switch from a left-handed spiral to a right-handed one. Massaging a kink to travel down the cord leads to a moving shape distortion, just like the motion that pushes Spiroplasma through liquid at speeds of about 3 to 5 micrometers, or about a body length, per second.
“The idea of this wave traveling down the length of the cell body is a simple one,” says Greg Huber, a physicist at the University of Connecticut Health Center in Farmington. “But Spiroplasma still has a lot to teach us.”
To explore the relationship between body design and swimming efficiency, Huber and his colleagues created a computer model that allowed the researchers to optimize different parameters, such as an organism’s cell length, the size of the kink-induced bend and how fast the kink travels down Spiroplasma’s body. “We’re trying to explore shapes that nature hasn’t,” Huber says.
The team’s results, which appeared online May 28 in Physical Review Letters, suggest that as with many things, nature nailed it: The model predicts values for an optimal Spiroplasma swimmer that are close to those of the real-life Spiroplasma. “What we find is that, just from those constraints alone, the optimal one is coincidentally the one nature has found! Is that a coincidence or is that evolution?” Huber says.
Artificial swimmers
Evolution’s success in hydrodynamic design at the microscale has attracted the attention of researchers interested in building small artificial swimmers. Such micromachines may one day carry chemotherapy drugs directly to the site of a tumor in the body, bust up plaques or clots in blood vessels, or take part in other small adventures hatched by creative scientists. But designing things that actually get around at such low Reynolds numbers isn’t easy.
“What’s new is to try to be audacious and think we can engineer something ourselves,” says Ramin Golestanian of the University of Sheffield in England. Designing tiny machines that swim, he says, is “a bit like the history of flying things. Birds with flapping wings are very smooth, but what we manage to make are these huge, ugly metallic things that fly — not very similar to the inspiration.”
A paper published online February 13 in Applied Physics Letters describes a bacteria-sized artificial swimmer. Mimicking corkscrew-shaped bacterial flagella, Li Zhang and his colleagues at the Institute of Robotics and Intelligent Systems in Zurich created tiny, magnetic flagella and attached them to a square-shaped metal head. These swimmers turn in one direction in response to a rotating magnetic field, propelling them through liquid at speeds around 1.2 micrometers per second. This is closer than previous attempts to match E. coli’s average speed, which approaches 30 μm/s.
Getting artificial swimmers out of the laboratory and swimming through human arteries or other real-world scenarios will be much harder, says Zhang, who led the new research. The magnetic swimmers were tested only in stationary liquid, he says, a much simpler feat than navigating the flowing liquid found almost everywhere microorganisms swim. “We have to find out how to steer them in a dynamic liquid environment and how to monitor them,” Zhang says.
Other researchers have taken a different approach to make artificial swimmers. Golestanian has modeled the chemical-powered, rocket booster–like propulsion of artificial particles in liquid. “We use chemical reactions, inspired by examples in nature,” he says. In a paper published May 8 in Physical Review Letters, Golestanian describes how fast certain kinds of these particles can move around and how they might be steered.
Some microorganisms can gravitate toward or away from certain chemicals, a process called chemotaxis. E. coli swimming toward a tasty meal of glucose is one such example. By building artificial swimmers that can detect and respond to varying concentrations of reactive chemicals in the surrounding liquid, researchers may be able to control the swimmers’ movements. “We can modulate the motion by making gradients or patterns — artificial chemotaxis, if you like — for these guys,” Golestanian says. “The exact form of the chemical reaction doesn’t matter. All that matters is that it’s fast enough.”
For now, such artificial microswimmers haven’t moved much beyond theoretical descriptions and rudimentary designs. But as researchers uncover more about liquid locomotion on tiny scales, the possibilities for such micromachines continue to expand. Says researcher Michael Graham of the University of Wisconsin–Madison: “It’s a … very fruitful area for combining ideas from physics and biology. It’s a complex problem, and that’s very enticing — lots of fertile ground.”







