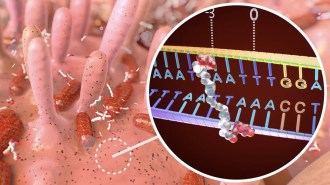
SMALL WORLD For their contributions to the development of cryo-electron microscopy, Jacques Dubochet, Joachim Frank and Richard Henderson (from left to right) won the 2017 Nobel Prize in chemistry.
From left: Axel Broquet, ©UNIL; Jorg Meyer; Medical Research Council
An imaging technique that freezes tiny biological objects such as proteins and viruses in place so that scientists can peer into their structures at the scale of atoms has won its developers the 2017 Nobel Prize in chemistry.
Jacques Dubochet of the University of Lausanne in Switzerland, Joachim Frank of Columbia University and Richard Henderson of the MRC Laboratory of Molecular Biology in Cambridge, England, won for their contributions to the development of the technique, called cryo-electron microscopy, the Royal Swedish Academy of Sciences announced October 4. The three will share the 9-million-Swedish-kronor (about $1.1 million) prize.
“Now that we can see the intricate details of every drop of our body fluids, we can understand how they are built and how they act and how they work together,” Sara Snogerup Linse, chair of the 2017 chemistry Nobel committee, said at a news conference. “We are facing a revolution in biochemistry…. Soon, there are no more secrets.”
Andrew Murray, a systems biologist at Harvard University, applauded the choice. “These are people whose work lies at the basis of an incredibly important technique.” The award, he says, “recognizes people who’ve done science for science’s sake…. The ultimate goal is to see molecules up close and personal.”
To map the minute landscape of molecules, at scales as tiny as just tenths of a nanometer, and help decipher their functions, structural biologists have long relied on two tools: nuclear magnetic resonance, or NMR, spectroscopy and X-ray crystallography. But both techniques have limitations. X-ray crystallography creates a 3-D picture of a molecule’s structure by bouncing X-rays off of a crystallized version of a molecule and reconstructing the molecule’s structure based on those diffraction patterns. “But a lot of proteins cannot be crystallized, so their structures are inaccessible to the prying mind of the biochemist,” Murray says. NMR, meanwhile, allows researchers to study biological molecules in a solution, such as water — but its use has generally been limited to relatively small proteins.
Enter cryo-electron microscopy, or cryo-EM. In the 1970s, Henderson first attempted to use regular electron microscopy to study a protein embedded in the membrane of a cell. He was unable to crystallize the protein and didn’t want to remove it from the membrane and thereby risk altering its structure.
Story continues after image
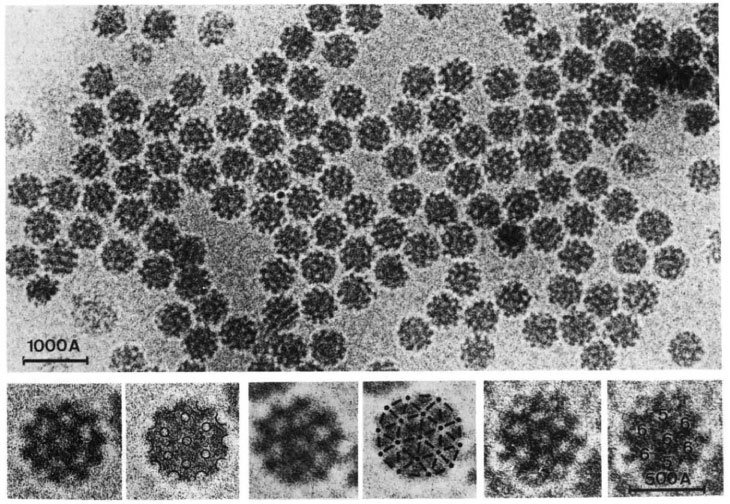
Electron microscopy wasn’t ideal for studying biological molecules. This technique works by shining a beam of electrons through a sample and observing how the electrons are deflected by the material they’ve passed through. The pattern of those deflections sketches out the material’s atomic structure. But the powerful electron beams can incinerate the material as they pass through it, weakening the beam and producing fuzzy images. Another drawback: The material must be in a vacuum in the microscope and that causes water within biological molecules to evaporate, distorting their shapes.
To get around these problems, in 1975 Henderson placed bacterial proteins, still within their membrane so that they retained their structures, within the microscope and covered the membrane with a glucose solution to keep it from drying out. He used a weaker electron beam to minimize the damage. Because the beam was weaker, the images were fuzzier than they would have been with a beam at full strength. But since the proteins in his sample happened to be oriented in the same direction, Henderson was able to compile the fuzzy images from each protein in the membrane to generate one sharper image. He then turned the membrane this way and that, capturing the proteins in different orientations and compiling those 2-D images to see the protein’s structure in three dimensions. The image, while an improvement over past efforts, wasn’t quite at the atomic scale yet — that required other advances.
One such advance came from Frank, who was tackling the issue of orientation. Unlike Henderson’s proteins, most proteins in a sample wouldn’t all be oriented in the same direction and conveniently ready to compile. Frank’s solution, published in 1981, was to create a computer algorithm to scan the images to look for any proteins oriented in the same direction and group them together. Like Henderson’s compiled image, each group in Frank’s image would be compiled to create a sharper picture.
Meanwhile, Dubochet was considering the problem of dehydration. Henderson’s glucose solution wouldn’t work for most biomolecules. Freezing samples also wouldn’t work, because ice crystals distort molecules’ shapes. But, he realized, if the water were to cool extremely rapidly to ‒196 ° Celsius — in ethane cooled by liquid nitrogen — it would solidify without forming ice crystals, instead becoming vitreous, or glassy. In 1984, he showed that viruses, when covered with a fine layer of vitrified water, could be safely studied in the electron microscope.
That cryo-EM technique dovetailed with Henderson’s efforts, and in 1990, Henderson produced the first cryo-EM image of a protein at the atomic scale. The following year, Frank prepared ribosomes, cellular structures that help make proteins, using the cryo-EM method. But for some time, the resolution of the images remained unsatisfying enough that many researchers called it “blobology.”
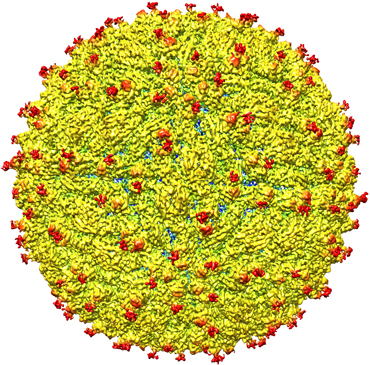
“Now, we need to apply this to look at larger complexes to understand how these different molecular machines work,” Subramaniam says.
Over the last few years, cryo-EM has begun to demonstrate its potential, including revealing the structure of larger and larger objects, such as viruses. In 2016, cryo-EM was used to map the structure of the Zika virus (SN: 4/30/16, p. 10), helping to identify possible regions to target with a vaccine or antiviral compounds.
“Cryo-EM has revolutionized structural biology, particularly in the last three years, with the invention of new kinds of electron detectors for the microscope,” says Michael Rossmann, a physicist and microbiologist at Purdue University in West Lafayette, Ind., and a coauthor of the Zika mapping study. “We call it the ‘resolution revolution.’” Next up, he says: looking at an entire cell.
Notes Subramaniam: “This is just the beginning. There are exciting years ahead.”